Sequence analysis of Epstein-Barr virus RPMS1 gene in malignant hematopathy of Northern China
Abstract
The RPMS1 gene is the only member of the BamHI-A rightward transcripts (BARTs) family for which a full-length complementary DNA has been identified, and RPMS1 transcript has been confirmed in many Epstein–Barr virus (EBV)-positive malignancies. However, the effects of sequence variations of RPMS1 in hematological malignancies and their biological significance are unclear. To explore the association between RPMS1 gene variations and hematological malignancy, the RPMS1 gene of 391 EBV-positive samples from patients with EBV-positive leukemia, myelodysplastic syndromes and lymphoma in northern China were sequenced. On the basis of phylogenetic tree and mutation characteristics of RPMS1, all the sequences were divided into five major types: RPMS1-A, RPMS1-B, RPMS1-C, RPMS1-E, and RPMS1-F. RPMS1-A type, similar to the prototype B95-8, was identified in 71.87% (281/391) of samples and was the major type in all subpopulations. The frequency of RPMS1-F type was significantly higher in all malignant hematopathy groups than in healthy donors. The Hodgkin lymphoma group contained more RPMS1-F than other malignant hematopathy groups, and acute myeloid leukemia contained more RPMS1-C type than other malignant hematopathy groups. Therefore, RPMS1-A is the main type of RPMS1 gene in northern China, and RPMS1-F may be associated with hematologic malignancies.
1 INTRODUCTION
Epstein–Barr virus (EBV) is a member of the herpesvirus family of lymphoblastic viruses. EBV infection is very common, and it has been shown that more than 90% of healthy adults worldwide carry the virus.1 EBV infection is closely related to the occurrence and development of various human cancers, including nasopharyngeal carcinoma (NPC), gastric carcinoma (GC), and multiple lymphomas, such as Burkitt lymphoma (BL), Hodgkin lymphoma (HL), and non-Hodgkin lymphoma (NHL).2-8 In addition to lymphomas, the relationship between EBV and other hematological malignancies has been gradually known in recent years, such as myelodysplastic syndromes (MDS), and leukemia.9, 10 Ferrajoli et al.9 have shown that EBV infection may be a factor in the progression of chronic lymphocytic leukemia.9 Borze et al.6 found that EBV-miR-BART13 was upregulated in all examined cases of MDS.10 In addition, they reported for the first time that active herpesviruses, especially EBV, exist in MDS patients, which provides evidence for the ability of MDS matrix factors to support viral activation.9
In addition to Epstein–Barr nuclear antigen (EBNA) family members, latent membrane proteins, EBV-encoded small RNAs, BamHI-A rightward transcripts (BARTs) are also expressed during latent EBV infection.11-15 Among these, the BARTs are the most widely expressed transcripts in latent infection of EBV, and they participate in the development of various EBV-related cancers. The RPMS1 gene is the only member of the BARTs family for which a full-length complementary DNA has been identified.16-18 Recombinant expression experiments showed that RPMS1 encodes 103 amino acids in bacteria, and biochemical activities of artificially expressed RPMS1 protein could be relevant to the role of the virus in cancer.18, 19 Feng et al.20 found that a single-nucleotide polymorphism (SNP) (locus 155391: G>A, named G155391A) in the second exon region of RPMS1 gene (located at 155267-155552 of the EBV genome) was significantly associated with the incidence of NPC. On this basis, our laboratory detected RPMS1 polymorphisms in EBV-related tumors (GC, NPC, and lymphoma).21 However, the relationship of RPMS1 variants with hematological malignancies and their biological significance are unclear.
In this study, the polymorphisms of RPMS1 gene were analyzed in patients with EBV-positive leukemia, MDS and lymphoma in North China. The intention is to investigate the roles of RPMS1 variants that may play in the development of hematologic malignancies.
2 MATERIALS AND METHODS
2.1 Samples and DNA extraction
A total of 798 peripheral blood samples were collected from patients in the Affiliated Hospital of Qingdao University, including 451 cases of leukemia, 172 cases of MDS, and 268 cases of lymphoma. Ninty-three throat washing (TW) samples were collected from healthy adults. Leukemia included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and chronic leukemia (CL), while lymphoma included HL and NHL. All the patients and healthy controls were inhabitants from Northern China. This study was approved by the Medical Ethics Committee of Qingdao University Medical College. Informed consent from all the study participants was received.
DNA was extracted and purified from TW, blood samples using standard protocols for proteinase K digestion and phenol-chloroform purification. All DNA extractions were stored at −20°C.
2.2 Determination of EBV-positive samples
The amplification of specific BamHI-W sequences in the EBV genome was detected by polymerase chain reaction (PCR). The primers used were as follows: forward, 5-CCCAACACTCCACCACACC-3; reverse, 5-TCTTAGGAGCTGTCCGAGGG-3. The PCR reactions were performed in a total volume of 25 μl containing 1× PCR reaction buffer, 200 μM of each deoxyribonucleotide triphosphate, 0.4 μM of each primer and 1 U Taq polymerase. The cycle conditions were as follows: predenaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s and final elongation at 72°C for 10 min.
2.3 PCR amplification of RPMS1
We amplified specific regions of RPMS1 using nested-PCR. For RPMS1, only the second coding exon was considered (sequence length approximately 282 bp, covering 89.74% of the RPMS1 coding region), as there was no variation in the first exon according to pairwise comparisons among GD1, AG876, and two wild-type EBV genomes (GenBank Accession No. AY961628, DQ279927, AJ507799, and NC_007605).20 The external and internal RPMS1 PCR primers are listed in Table 1. RPMS1-1 and RPMS1-2 were used for the first round of amplification, whereas RPMS1-3 and RPMS1-4 were used for the second round of amplification. The PCR reactions were performed in a total volume of 30 μl containing 1× PCR reaction buffer, 200 μM of each deoxyribonucleotide triphosphate, 0.4 μM of each primer and 1 U Taq polymerase. The cycle conditions were as follows: predenaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 1 min and final elongation at 72°C for 10 min. A water control, negative control and positive control were set up for each PCR. The EBV-positive cell line B95-8 was used as a positive control, and EBV-negative cell line (Jurkat) was used as a negative control.
| Name of primers | Sequence (5′–3′) | B95-8 coordinates | Size of PCR products |
|---|---|---|---|
| RPMS1-1 | GCTGGGTTGATGCTGTAGATG | 155088–155109 | 712 bp |
| RPMS1-2 | AGGGTCTGGACGTGGAGTTTG | 155821–155800 | |
| RPMS1-3 | AGATGTGCCTGGCTCTGTC | 155104–155123 | 440 bp |
| RPMS1-4 | CAATGACTTTGTCACCTTTGG | 155565–155544 |
2.4 Sequencing analysis of PCR products
We used 25 μl of the amplification products from the second round of PCR for bidirectional sequencing using the end-termination method (BGI), and the sequencing primers were RPMS1-3 and RPMS1-4, as shown in Table 1. Peak map files were used to check the sequencing results with Chromes software. DNA Star software (Lasergene, version7.0) was used to analyze splicing and alignment. The EBV standard strain B95-8 (GenBank number, NC_007605) was used as the reference sequence and was compared with the sequencing results we obtained.
2.5 Statistical analysis
The χ2 and Fisher's exact tests were used to analyze the distributions of RPMS1 variants between different population groups. Statistical analysis was carried out using SPSS15.0 statistical software (SPSS), and differences with values less than 0.05 (p < 0.05) were considered significant.
3 RESULTS
3.1 EBV infection in patients with malignant hematological diseases
A total of 307 EBV-positive malignant hematological specimens were detected, including 137 leukemia specimens, 56 MDS specimens, and 114 lymphoma specimens. The positive rates infected with EBV in each group are shown in Table 2.
| RPMS1 | Leukemia | MDSs | lymphoma, | TWs |
|---|---|---|---|---|
| subtypes | n (%) | n (%) | n (%) | n (%) |
| RPMS1-A | 94 (68.6) | 44 (78.57) | 74 (64.9) | 69 (82.1) |
| RPMS1-B2 | 7 (5.1) | 2 (3.57) | 4 (3.5) | 12 (14.3) |
| RPMS1-C | 14 (10.2) | 1 (1.79) | 9 (7.9) | 2 (2.4) |
| RPMS1-E | 6 | 1 | 4 | 0 |
| RPMS1-F | 16 | 8 | 38 | 1 |
| Total | 137 (100) | 56 (100) | 114 (100) | 84 (100) |
- Abbreviations: EBV, Epstein–Barr virus; MDS, myelodysplastic syndromes; TW, throat washings from healthy donors.
3.2 Sequence variations of RPMS1
A total of 391 EBV-positive samples were successfully sequenced for the RPMS1 gene, including 46 ALL cases, 50 AML cases, 41 CL cases, 56 MDS cases, 18 HL cases, 96 NHL cases, and 84 TWs of healthy donors.
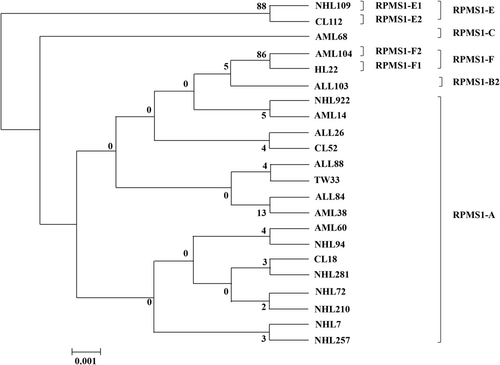
No double peaks were detected at single-nucleotide positions in any of the specimens, indicating that all were single RPMS1 sequences. Compared with the RPMS1 sequence of the B95-8 standard strain, 22 nucleotide mutations were found in RPMS1 across all the samples, including 18 missense mutations and 4 synonymous mutations, as shown in Figure 1. According to the phylogenetic tree, the EBV isolates belonged to five major types (RPMS1-A, RPMS1-B, RPMS1-C, RPMS1-E, and RPMS1-F) and could be further divided into seven subtypes (RPMS1-A, RPMS1-B2, RPMS1-C, RPMS1-E1, RPMS1-E2, RPMS1-F1, and RPMS1-F2), as shown in Figure 2.
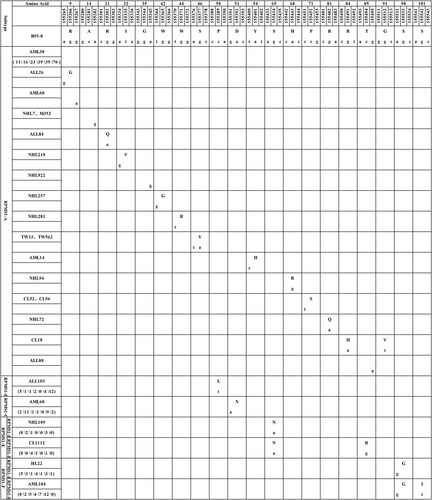
A total of 281 specimens with amino acid sequences similar to B95-8 were identified, including 34 ALL, 32 AML, 28 CL, 44 MDS, 10 HL, 64 NHL, and 69 TW samples from healthy donors. Four synonymous mutations (g155267a, a155282g, a155345g, and g155495a) were, respectively, found in 5 specimens. In addition, sporadic amino acid mutations (R9G, R21Q, I32V, W42G, W44R, S46Y, Y54H, H68R, P71S, R81Q, R84H, and G91V) were detected in 13 specimens. The above isolates with the same RPMS1 amino acid sequence as B95-8 or sporadic mutations were classified as type RPMS1-A.
We found that 25 specimens had a P50L mutation at the 50th codon, including 5 AML, 1 ALL, 1 CL, 2 MDS, 4 NHL, and 12 TW samples. In addition, D51N mutations were found in 26 specimens, including 2 AML, 11 ALL, 1 CL, 1 MDS, 9 NHL, and 2 TW samples. According to Wu et al., we respectively classify these two mutations into groups RPMS1-B2 and RPMS1-C.21
Eleven specimens had an S65N mutation at the 65th codon, including 1 AML, 5 CL, 1 MDS, and 4 NHL samples, in which 6 specimens simultaneously had another mutation, T85R. These two sequences were classified as subtypes RPMS1-E1 and RPMS1-E2, respectively, and they belonged to RPMS1-E type. There were 18 specimens with S98G mutations at codon 98 separately, and these isolates were classified as the RPMS1-F1 subtype. S98G and S101I mutations were simultaneously detected in 30 specimens. We classified these isolates as RPMS1-F2 subtype. Subtypes RPMS1-F1 and RPMS1-F2 together constituted the RPMS1-F type.
3.3 Phylogenetic tree analysis
The phylogenetic tree constructed from the 22 RPMS1 representative sequences is shown in Figure 2. The phylogenetic tree has seven branch lines in accordance with seven subtypes, RPMS1-A, RPMS1-B2, RPMS1-C, RPMS1-E1, RPMS1-E2, RPMS1-F1, and RPMS1-F2.
3.4 Distribution of RPMS1 types in different diseases and healthy donors
The distribution of RPMS1 gene types in leukemia, MDS, and lymphoma patients and in healthy humans from North China is shown in Table 2. RPMS1-A was the predominant type, accounting for 71.87% of samples (281/391). The frequency of RPMS1-A in TW samples was significantly higher than that in leukemia and lymphoma (TW vs. leukemia, χ2 = 4.924, p = 0.026; TW vs. lymphoma, χ2 = 7.157, p = 0.007). There were no significant differences between TW and MDS (p > 0.05).
The frequency of RPMS1-B2 subtype in TW samples was also significantly higher than those in leukemia, MDS, and lymphoma (TW vs. leukemia, χ2 = 5.580, p = 0.018; TW vs. MDS, χ2 = 4.286, p = 0.038; TW vs. lymphoma, χ2 = 7.562, p = 0.006). The frequency of the RPMS1-C type in TW samples was significantly lower than that of leukemia (TW vs. leukemia, χ2 = 13.758, p < 0.001), but there was no significant difference with MDS and lymphoma (p > 0.05). The frequency of RPMS1-E in TW samples was significantly lower than in leukemia and lymphoma samples (TW vs. leukemia, χ2 = 5.84, p = 0.005; TW vs. lymphoma, χ2 = 4.477, p = 0.034), but not significantly different from MDS samples. The frequency of RPMS1-F type in TW samples was significantly lower than that in leukemia, MDS, and lymphoma (TW vs. leukemia, χ2 = 8.067, p = 0.005; TW vs. MDS, χ2 = 10.025, p = 0.002; TW vs. lymphoma, χ2 = 16.365, p < 0.001).
3.5 Distribution of RPMS1 types in different hematologic malignancies
The distribution of RPMS1 gene types in different hematologic malignancies is shown in Table 3. The frequency of the RPMS1-C type was significantly higher in leukemia samples than in MDS samples (leukemia vs. MDS, χ2 = 5.025, p = 0.025), among them, the frequency of RPMS1-C in AML samples was significantly higher than that in ALL, CL, MDS, HL, and NHL samples (AML vs. ALL, χ2 = 6.376, p = 0.012; AML vs. CL, χ2 = 7.530, p = 0.006; AML vs. MDS, χ2 = 10.752, p = 0.001; AML vs. HL, χ2 = 4.724, p = 0.030; AML vs. NHL, χ2 = 4.433, p = 0.035). Although there was no significant difference between the frequency of RPMS1-C in lymphoma samples and MDS samples, the frequency of RPMS1-C in NHL samples was significantly higher than in MDS samples (NHL vs. MDS, χ2 = 3.984, p = 0.046).
| RPMS1 | ALLs | AMLs | CLs | MDSs | HLs | NHLs |
|---|---|---|---|---|---|---|
| subtypes | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| RPMS1-A | 34 (73.9) | 32 (64.0) | 28 (68.4) | 44 (78.57) | 10 (55.6) | 64 (66.7) |
| RPMS1-B2 | 5 (10.9) | 1 (2.0) | 1 (2.4) | 2 (3.57) | 0 | 4 (4.2) |
| RPMS1-C | 2 (4.3) | 11 (22.0) | 1 (2.4) | 1 (1.79) | 0 | 9 (9.4) |
| RPMS1-E | 0 | 1 (2.0) | 5 (12.2) | 1 (1.79) | 0 | 4 (4.2) |
| RPMS1-F | ||||||
| RPMS1-F1 | 5 (10.9) | 3 (6.0) | 1 (2.4) | 4 (7.14) | 1 (5.5) | 3 (3.1) |
| RPMS1-F2 | 0 | 2 (4.0) | 5 (12.2) | 4 (7.14) | 7 (38.9) | 12 (12.5) |
| Total | 46 (100) | 50 (100) | 41 (100) | 56 (100) | 18 (100) | 96 (100) |
- Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CL, chronic leukemia; HL, Hodgkin's lymphoma; MDS, myelodysplastic syndromes; NHL, non-Hodgkin's lymphoma.
In addition, although the frequency of RPMS1-E in leukemia had no significant difference with MDS samples, the frequency of RPMS1-E in CL samples was significantly higher than that in ALL, AML, MDS and lymphoma samples (CL vs. ALL, χ2 = 7.867, p = 0.005; CL vs. AML, χ2 = 4.016, p = 0.045; CL vs. MDS, χ2 = 4.578, p = 0.032; CL vs. lymphoma, χ2 = 4.16, p = 0.041). And we found in detailed study that the frequency of RPMS1-F was significantly higher in HL samples than in ALL, AML, CL, MDS, and NHL samples (HL vs. ALL, χ2 = 8.244, p = 0.004; HL vs. AML, χ2 = 9.119, p = 0.003; HL vs. CL, χ2 = 5.788, p = 0.016; HL vs. MDS, χ2 = 6.604, p = 0.01; HL vs. NHL, χ2 = 6.701, p = 0.01).
4 DISCUSSION
EBV may play a role in the pathogenesis of leukemia, MDS, and lymphomas.6, 8, 10, 22, 23 In our studies, EBV-DNA were found in 137 of the 451 (30.4%) leukemia samples, 56 of the 172 MDS samples (32.6%) and in 114 of the 268 lymphomas samples (42.5%). The frequencies of EBV infection, as described here, were similar to those reported in AL patients (34.6%).23 In summary, EBV may play an important role in the pathogenesis of leukemia, MDS, and lymphomas. Therefore, it is important to better understand the relationship between EBV infection and hematological malignancies.
The BART family represents an important pathogenic gene of EBV, encodes complex gene products, and is stably expressed in all types of infected cells. Recombinant expression experiments showed that RPMS1 encodes 103 amino acids in bacteria, and hybridization experiments showed that RPMS1 could interact with RBP-Jk/CBF1.18 Biochemical activities of artificially expressed RPMS1 protein have been identified and could be relevant to the role of the virus in cancer.19 In addition, RPMS1 was found to antagonize transcription activation by Notch 1 and EBNA2 by competing for binding to RBP-Jk.24 However, no endogenous RPMS1 protein has been reported in cultured NPC cells or NPC tumor biopsies.25 Motivated by the above results, RPMS1 gene mutations were examined in this study.
In this study, RPMS1 gene was successfully sequenced and polymorphically analyzed. The results showed that EBV RPMS1 was highly conserved among hematological malignancies and the healthy population. The type A with similar amino acid sequence to the standard B95-8, was most prominent (accounting for 71.87% of isolates), which was in agreement with our previous study of other EBV-positive malignancies, including GC and NPC in the same area.21
To date, large studies have identified RPMS1 sequence variant associated with high risk of NPC represented by g155391a,20, 21, 26 while other studies have found that it may not be associated with other EBV-related tumors, such as EBV-associated GC, BL, and HL.21, 26 The correlation with NPC indicates that RPMS1 g155391a may be specific for the occurrence and development of NPC. Mutation of guanine to adenine in the RPMS1 g155391a variant results in aspartic acid becoming asparagine, which may be related to the transcription or expression of RPMS1. A related study showed that the frequency of SNP g155391a also known as RPMS1-C, was significantly higher in South China than that in North China and Europe.20 Correia et al.27 also found that RPMS1-C was more frequently found in NPC in South China. In our study, the frequency of RPMS1-C was significantly higher in the AML group than that in the other tumors groups, and the incidence of RPMS1-C in the NHL group was significantly higher than that in the MDS group. It is worth noting that this study showed that the RPMS1-C subtype g155391a was present in EBV-positive leukemia, whereas this variation was rare in EBV-positive MDS. MDS is a heterogeneous myeloid clonal disease originating from hematopoietic stem cells. It has a higher probability of transformation into AML and is a precursor of leukemia.28 Therefore, it was speculated that the 51st amino acid variation of RPMS1 may be related to the transformation of MDS into leukemia, but this proposed mechanism requires further investigation.
A study by Wu revealed three new polymorphisms (g155325t, g155326a, and c155389t) that had not been discovered before.21 However, in our experiment, only the c155389t mutation was found. Furthermore, we discovered four novel polymorphisms (g155434a, c155494g, a155532g, g155542t) in our experiment that had not been previously identified. These four missense mutations lead to S65N, T85R, S98G, and S101I translation mutations and were classified into E and F types, respectively. This study shows that there were two variants of RPMS1-E subtype g155434a and c155494g in EBV-positive acute leukemia, whereas this variation occurs more frequently in EBV-positive CL. Therefore, it was speculated that the 65st amino acid and 85st amino acid variations of RPMS1 may be related to the occurrence and development of CL. RPMS1-F occurred more frequently in all detected patients with hematological malignancies than in healthy adults, so we suspect that RPMS1-F is related to hematological malignancies. Feng et al.20 functionally characterized a site of SNP and found that this locus functionally regulates the protein stability of RPMS1. And the G to A mutation caused a longer half-life of RPMS1 protein, as shown in the protein degradation assays. These results support the speculation that RPMS1 may be converted to protein at very low levels or that RPMS1 protein is degraded very rapidly. Mutation of adenine to guanine and guanine to thymine in the RPMS1 a155532g and g155542t variant results in serine acid becoming glycine and serine acid becoming isoleucine, which may change the conformation or biological function of RPMS1 protein. The rate between the healthy group and other EBV-associated malignant hematological diseases of RPMS1-F indicates that RPMS1 SNP a155532g and g155542t may be associated with a higher risk of malignant hematological diseases. And the prevalence of RPMS1-F was significantly higher in HL than in other subgroups, including the NHL group. However, due to the insufficient data of the HL group, it is necessary to exaggerate the sample size for analysis.
In the present study, the polymorphism expression patterns of RPMS1 were significantly different between malignant hematological patients and healthy donors. The frequency of RPMS1-A was significantly higher in the samples from healthy donors. While substrains with concomitant mutations, especially RPMS1-F, were more common in hematological malignancies. This suggests that EBV RPMS1 polymorphism may be disease specific in hematologic malignancies. However, the exact relationship between EBV infection and malignant hematoma and its role in the disease need further study.
AUTHOR CONTRIBUTIONS
Bing Luo: conceived the project and designed experiments. Ping Li and Lei Liu: mainly obtained specimens from the Affliated Hospital of Qingdao University. Meng-He Zhao: mainly conducted the experiments, analyzed the data and wrote the draft of the manuscript. Xing Zhang and Wen Liu: participated in performance of the experiments and the drafting of the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENT
This research was supported by Natural Science Foundation of Shandong Province (ZR2020MH302).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENTS
This research was approved by the Medical Ethical Committee of Affiliated Hospital, Qingdao University (QYFY WZLL 26610) and has been granted an exemption from requiring written informed consent.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analysed during this study are included in this. Further enquiries can be directed to the corresponding author.




