Whole-transcriptome sequencing data reveals a disparate cognitive and immune signature in COVID-19 patients with and without dementia
Abstract
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused more than 6.3 million deaths worldwide. Recent evidence has indicated that elderly people with dementia are particularly vulnerable to COVID-19 and severe disease outcomes. However, its molecular mechanism remains largely unknown. Here, we retrieved frontal cortex samples of COVID-19 patients from the Gene Expression Omnibus database and performed a systematic transcriptomic analysis to compare COVID-19 patients and controls with or without dementia. In nondemented patients, SARS-CoV-2 infection obviously activated T helper type 2 (Th2) cell-mediated humoral immunity and reduced the pathogenesis of dementia-related Alzheimer's disease and Parkinson's disease. In demented patients, conversely, SARS-CoV-2 infection significantly increased T helper type 1 (Th1) cell-mediated cellular immunity and exacerbated the progression of dementia-related diseases. We further analyzed the molecular characteristics of COVID-19 patients with and without dementia. Compared with nondemented COVID-19 patients, demented COVID-19 patients showed decreased enrichment scores of Calcium signaling pathway, Neuroactive ligand-receptor interaction, ABC transporters, and Peroxisome, and increased enrichment scores of Olfactory transduction and Regulation of autophagy. The ratio of Th1/Th2 cells was significantly increased from 1.17 in nondemented COVID-19 patients to 33.32 in demented COVID-19 patients. Taken together, our findings provide transcriptomic evidence that COVID-19 has distinct influences on cognitive function and immune response in patients with and without dementia.
1 INTRODUCTION
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has rapidly spread to become a pandemic. According to the World Health Organization (WHO), as of June 30, 2022, more than 543.3 million cases and 6.3 million deaths have occurred worldwide.1 Elderly people are considered more susceptible to COVID-19 and have higher mortality than any younger age group.2-5 Limited data suggest that SARS-CoV-2 can enter the central nervous system and preferentially target the frontal lobes.6-8 For both vaccinated and unvaccinated patients, pre-existing dementia increased the risk of COVID-19-related hospitalization and death by more than twofold.9-11 Hence, it is urgent to understand the molecular mechanism underlying high mortality caused by COVID-19 in demented patients.
Dementia is a syndrome characterized by progressive cognitive impairment and ranks as a leading cause of disability among older people worldwide.12 According to estimates from the WHO, the number of people living with dementia is expected to double over the next decades, from 78 million in 2030 to 139 million in 2050.13 Alzheimer's disease is the most common cause of dementia, accounting for an estimated 60%–80% of cases. Other common causes of dementia include cerebrovascular disease, frontotemporal lobar degeneration, and Parkinson's disease.14 Extensive studies have provided clear evidence that dementia significantly increases the SARS-CoV-2 infection risk and COVID-19-related mortality, indicating heterogeneity and a distinct molecular pattern between patients with and without dementia.15-17 However, few studies have paid full attention to the difference between the demented and nondemented cases during COVID-19.
To explore the possible difference between the demented and nondemented patients with COVID-19 and the potential mechanisms, we retrieved two public cohort datasets from the Gene Expression Omnibus (GEO) database, and characterized the transcriptomic changes in the frontal cortex tissues of COVID-19 patients and controls with or without dementia. Our data revealed a disparate influence of COVID-19 on cognitive function and immune response between patients with and without dementia and provided a possible link between COVID-19 and dementia by regulating the inflammation.
2 MATERIALS AND METHODS
2.1 Data source
Two COVID-19-related human frontal cortex expression profiling datasets, GSE188847 and GSE164332, were downloaded from the GEO database. GSE188847 data set comprised 12 COVID-19 patients and 12 age-matched controls (mean age 66.6 years old), with no known psychiatric or neurological disorders (platform: GPL24676; Illumina NovaSeq 6000).18 GSE164332 data set included eight COVID-19 patients (six patients with dementia and two patients without dementia) and 6 controls with dementia (mean age 83.8 years old) (platform: GPL18573; Illumina NextSeq 500), and the dementia status was evaluated by a forensic medical doctor and a neurologist.19 The clinical characteristics of patients used in this study were listed as Table 1.
| GEO data set | GEO accession | Disease type | Dementia status | Age | Sex | Country |
|---|---|---|---|---|---|---|
| GSE188847 | GSM5690952 | COVID-19 | No dementia | 46 | Male | United States |
| GSM5690953 | COVID-19 | No dementia | 57 | Female | United States | |
| GSM5690954 | COVID-19 | No dementia | 58 | Female | United States | |
| GSM5690955 | COVID-19 | No dementia | 62 | Male | United States | |
| GSM5690956 | COVID-19 | No dementia | 62 | Male | United States | |
| GSM5690957 | COVID-19 | No dementia | 64 | Male | United States | |
| GSM5690958 | COVID-19 | No dementia | 65 | Male | United States | |
| GSM5690959 | COVID-19 | No dementia | 72 | Female | United States | |
| GSM5690960 | COVID-19 | No dementia | 75 | Female | United States | |
| GSM5690961 | COVID-19 | No dementia | 75 | Male | United States | |
| GSM5690962 | COVID-19 | No dementia | 80 | Female | United States | |
| GSM5690963 | COVID-19 | No dementia | 84 | Male | United States | |
| GSM5690964 | Control | No dementia | 45 | Male | United States | |
| GSM5690965 | Control | No dementia | 59 | Female | United States | |
| GSM5690966 | Control | No dementia | 59 | Female | United States | |
| GSM5690967 | Control | No dementia | 61 | Male | United States | |
| GSM5690968 | Control | No dementia | 62 | Male | United States | |
| GSM5690969 | Control | No dementia | 64 | Male | United States | |
| GSM5690970 | Control | No dementia | 64 | Male | United States | |
| GSM5690971 | Control | No dementia | 71 | Female | United States | |
| GSM5690972 | Control | No dementia | 75 | Female | United States | |
| GSM5690973 | Control | No dementia | 75 | Male | United States | |
| GSM5690974 | Control | No dementia | 80 | Female | United States | |
| GSM5690975 | Control | No dementia | 84 | Male | United States | |
| GSE164332 | GSM5006430 | Control | Dementia | 78 | Female | Italy |
| GSM5006432 | Control | Dementia | 84 | Male | Italy | |
| GSM5006433 | Control | Dementia | 85 | Female | Italy | |
| GSM5006434 | Control | Dementia | 80 | Male | Italy | |
| GSM5006435 | Control | Dementia | 104 | Female | Italy | |
| GSM5006436 | Control | Dementia | 84 | Female | Italy | |
| GSM5006437 | COVID-19 | Dementia | 74 | Female | Italy | |
| GSM5006438 | COVID-19 | Dementia | 87 | Male | Italy | |
| GSM5006439 | COVID-19 | No dementia | 67 | Male | Italy | |
| GSM5006440 | COVID-19 | Dementia | 94 | Female | Italy | |
| GSM5006442 | COVID-19 | No dementia | 80 | Female | Italy | |
| GSM5006443 | COVID-19 | Dementia | 83 | Female | Italy | |
| GSM5006444 | COVID-19 | Dementia | 92 | Male | Italy | |
| GSM5006445 | COVID-19 | Dementia | 81 | Male | Italy |
2.2 RNA-seq analysis
Raw RNA-seq data were downloaded from the GEO database using the ARCHS4 tool and aligned against the GRCh38 human reference genome; the transcript counts were mapped to the gene level using human_matrix_v10 using R software (version 4.2.0).20 Differentially expressed genes (DEGs) were analyzed using NetworkAnalyst 3.0.21 Genes with a variance percentile rank lower than 15 and count lower than 4, and unannotated genes were removed. Normalization was performed by log2-counts per million transformation. The DESeq2 algorithm was used to identify DEGs according to the criteria |log2 fold change| > 0.6 and p < 0.05.
2.3 Functional enrichment analysis and gene set enrichment analysis (GSEA)
Process enrichment analysis of DEGs was conducted with Gene Ontology (GO) Biological Processes sources using Metascape (https://metascape.org).22 Terms with a p < 0.05 were considered significantly enriched. Crucial pathways involved in COVID-19 were analyzed with the gene set database c2.cp.kegg.v7.5.1.symbols.gmt and 1000 permutations using GSEA 4.2.3 software.23 According to the instructions, if the sample size of each phenotype was more than 7, the permutation type chose phenotype; if the sample size of each group was more than 3, the permutation type chose gene_set and the genes were ranked by the Signal2Noise. Each phenotype has less than three samples, the permutation type chose gene_set and the genes were ranked by the Diff_of_Classes. Pathways with nominal p < 0.05 and |Normalized Enrichment Score| > 1 were considered statistically significant.
2.4 Protein–protein interaction (PPI) network construction and hub gene analysis
The PPI network of DEGs was constructed using STRING v11.5 and the interaction score was set as medium confidence (0.400).24 Then the network was visualized using Cytoscape v3.8.2.25 The centralities of each node were calculated using CentiScaPe 2.2 plug-in, and the nodes were ranked based on degree.26 Genes with the top 20 degree values were identified as hub genes.
2.5 Immune cell infiltration analysis
xCell (https://xcell.ucsf.edu/) is a webtool that performs cell type enrichment analysis of 36 immune cell types based on gene signatures using bulk gene expression data.27 The gene-level read counts were normalized to transcripts per million and then used to estimate the effects of COVID-19 on the brain infiltrating immune cell subsets and abundance using xCell.
2.6 Statistical analysis
Data were expressed as the means ± standard deviation. Differences between groups were analyzed for significance by the unpaired t-test using GraphPad Prism 9 (GraphPad Software Inc.). Pearson's correlation was calculated to verify the relationship between hub genes and dementia-related biomarker genes (APP, MAPT, and SNCA) on the basis of the normalized expression values. A p-value less than 0.05 was defined as statistically significant.
3 RESULTS
3.1 COVID-19 enhanced the Th2 cell-mediated humoral immune response and reduced dementia pathogenesis in patients without dementia
We first determined the effects of COVID-19 on the gene transcription levels in the frontal cortex of the human brain from GEO GSE188847. As shown in Figure 1A, A total of 1507 differentially expressed genes were identified according to the criteria |log2 fold change| > 0.6 and p < 0.05, including 989 upregulated and 518 downregulated DEGs. Biological process enrichment analysis revealed that the 989 upregulated DEGs were enriched in immune-related processes, such as inflammatory response, cellular response to cytokine stimulus, positive regulation of immune response, cytokine production, and immune system process, and were also strongly related to the regulation of defense response, positive regulation of cell death and response to virus (Figure 1B). Meanwhile, the 518 downregulated DEGs were mainly distributed in nervous system-related processes, such as synaptic signaling, chemical synaptic transmission, neuropeptide signaling pathway, synapse organization, and synapse structure or activity, indicating that COVID-19 might affect patients' behavior and brain function.
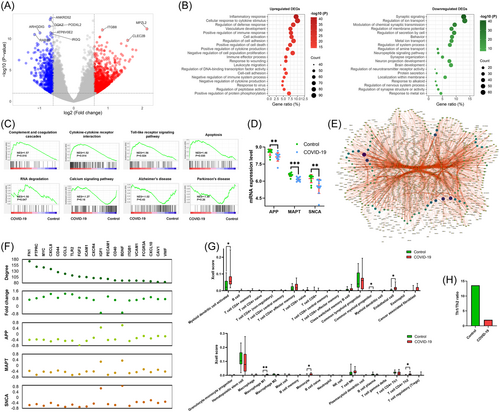
As shown in Figure 1C, the GSEA results demonstrated that COVID-19 obviously increased the enrichment scores of immune-related pathways, such as Complement and coagulation cascades (p = 0.016), Cytokine–cytokine receptor interaction (p = 0.014), Toll-like receptor signaling pathway (p = 0.024), as well as Apoptosis (p = 0.045) and RNA degradation (p = 0.047). Conversely, patients with COVID-19 showed decreased enrichment scores of Calcium signaling pathway (p = 0.12), Alzheimer's disease (p = 0.428), and Parkinson's disease (p = 0.257) compared with patients in the control group.
Strikingly, COVID-19 significantly decreased the levels of dementia-related biomarker genes APP, MAPT, and SNCA (Figure 1D). Then, we constructed a PPI network and identified 20 hub genes from the network (Figure 1E). To further verify the involvement of COVID-19 and dementia, we performed a correlation analysis between hub genes and dementia-related biomarkers. As shown in Figure 1F, expression levels of 20 hub genes were significantly changed in patients with COVID-19. The 18 upregulated hub genes in COVID-19 negatively correlated with dementia-related biomarkers, while 2 downregulated hub genes positively correlated with dementia-related biomarkers.
As aging is an important risk factor for dementia, we divided the patients into younger and older groups based on the mean age and explored the potential effects of COVID-19 in different age groups.28-30 GSEA results revealed that COVID-19 increased the enrichment scores of immune-related pathways and reduced the enrichment scores of Alzheimer's disease and Parkinson's disease in both younger and older groups (Supporting Information: Figure 1A,C). However, this tendency in the elder group was more obvious than that of the younger group. A decreased expression levels of APP, MAPT, and SNCA were observed in patients with COVID-19 in both younger and older groups (Supporting Information: Figure 1B,D).
To confirm the association of COVID-19 and the immune response, we analyzed the immune cell infiltration in frontal cortex tissues. Compared with the control group, COVID-19 patients exhibited higher levels of myeloid dendritic cell activated, endothelial cell, macrophage M1, monocyte and T cell CD4 + Th2, but a lower level of common myeloid progenitor (Figure 1G). In addition, the ratio of Th1/Th2 cells was dramatically decreased from 13.58 in the control group to 2.01 in the COVID-19 group (Figure 1H). These findings indicate that the host immune system might protect the nervous system from SARS-CoV-2 infection by inhibiting the Th1 cell-mediated cellular immune response.
3.2 COVID-19 decreased the Th2 cell-mediated humoral immune response and accelerated dementia pathogenesis in patients with dementia
Next, we assessed the influence of COVID-19 in patients with dementia. We extracted 803 DEGs, including 333 upregulated and 470 downregulated DEGs (Figure 2A). Functional enrichment analysis showed that the 333 upregulated DEGs were closely associated with cytoplasmic translation, ribosomal small subunit biogenesis, mRNA processing, cellular component biogenesis, and regulation of synaptic transmission, dopaminergic (Figure 2B). The 470 downregulated DEGs were markedly correlated with cellular response to cytokine stimulus, response to wounding, inflammatory response, and cellular homeostasis.
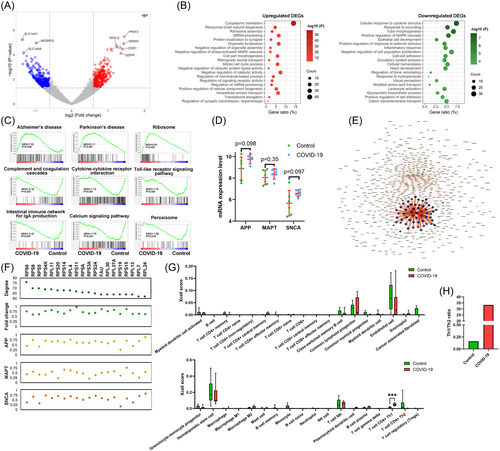
As shown in Figure 2C, GSEA confirmed that COVID-19 was positively correlated with the pathways Alzheimer's disease (p = 0.004), Parkinson's disease (p < 0.001), and Ribosome (p < 0.001), but was negatively interrelated with the pathways Complement and coagulation cascades (p < 0.001), Cytokine–cytokine receptor interaction (p < 0.001), Toll-like receptor signaling pathway (p < 0.001), Intestinal immune network for IgA production (p < 0.001), Calcium signaling pathway (p < 0.001), and Peroxisome (p = 0.018).
As shown in Figure 2D, COVID-19 slightly increased levels of APP, MAPT, and SNCA. From the PPI network, we identified 20 hub genes (Figure 2E). Furthermore, higher levels of the 20 hub genes were observed in demented COVID-19 patients than in demented controls (Figure 2F). All 20 hub genes were positively associated with dementia-related biomarker genes.
According to the mean age, patients in GSE164332 data set were divided into younger and older groups. Compared with controls, the enrichment scores of Alzheimer's disease and Parkinson's disease pathways were increased, while the immune-related pathways were downregulated in patients with COVID-19 in both younger and older groups (Supporting Information: Figure 2A,C. Meanwhile, the enrichment scores of immune-related pathways were decreased in patients with COVID-19 in both groups, except Cytokine–cytokine receptor interaction pathway in the younger group. As shown in Supporting Information: Figure 2B,D, patients with COVID-19 exhibited high expression levels of APP, MAPT, and SNCA in both groups.
Xcell analysis revealed that the infiltration level of T cell CD4 + Th1 was significantly upregulated in demented COVID-19 patients (p < 0.001, Figure 2G). In addition, patients with COVID-19 showed a sharp increase of Th1/Th2 ratio, from 0.16 to 33.32 (Figure 2H). The results suggest that COVID-19 might impair the nervous system and promote the progression of dementia-related diseases by activating Th1 cell-mediated cellular immunity.
3.3 Molecular signatures of COVID-19 patients with and without dementia
To clarify the different responses of patients with and without dementia during COVID-19, we analyzed the expression profiles of frontal cortex tissues from 6 demented and 2 nondemented patients who were infected with SARS-CoV-2. As shown in Figure 3A, 196 DEGs were obtained, including 79 upregulated and 117 downregulated DEGs. The 79 upregulated DEGs showed a significant association with mitogen-activated protein kinase (MAPK) cascade, endothelial cell migration, cellular response to acid chemical, lipoprotein metabolic process, and cellular response to hypoxia (Figure 3B). The 117 downregulated DEGs were obviously enriched in several immune-related biological processes, such as stimulatory C-type lectin receptor signaling pathway, inflammatory response, positive regulation of leukocyte cell-cell adhesion, lymphocyte-mediated immunity, and leukocyte differentiation.

The GSEA results demonstrated that Calcium signaling pathway (p < 0.001), Neuroactive ligand-receptor interaction (p < 0.001), Leishmania infection (p < 0.001), ABC transporters (p = 0.003), and Peroxisome (p = 0.026) were enriched in patients without dementia. However, the enrichment degree of Olfactory transduction (p < 0.001), Ribosome (p = 0.10), Regulation of autophagy (p = 0.060), Porphyrin and chlorophyll metabolism (p = 0.079) was significantly higher in demented patients than in nondemented patients (Figure 3C). During COVID-19, demented patients exhibited less B cell infiltration, and a higher level of T cell CD4 + Th1 than nondemented patients (Figure 3D). The ratio of Th1/Th2 was significantly increased from 1.17 in nondemented patients to 33.32 in demented patients (Figure 3E).
4 DISCUSSION
In the present study, we found that COVID-19 was strongly associated with nervous system processes in both demented and nondemented, but the effects of COVID-19 on the DEG expression profile differed between the groups. In nondemented patients, COVID-19-downregulated DEGs were related to synaptic function and neuropeptide signaling pathway. In demented patients, COVID-19-upregulated DEGs were related to synaptic function. The GSEA results also revealed an unequivocal role of COVID-19 in the progression of dementia-related diseases. COVID-19 could slightly attenuate the risk of Alzheimer's disease and Parkinson's disease in nondemented patients. Meanwhile, COVID-19 significantly increased the risk of dementia in demented patients. Although aging is a major risk factor for cognitive deficits and neurodegenerative diseases, we observed the same pattern of COVID-19 on immune response and dementia in the younger and elder groups. One study reported that COVID-19 can cause cognitive decline in young patients.31 This does not seem to conflict with our conclusion because even for healthy people, long-term isolation and hospitalization can also cause a noticeable impact on mental state.
Subsequently, we identified 20 hub genes and performed a correlation analysis of the expression levels of the 20 hub genes and 3 dementia-related biomarkers. APP and MAPT genes encode amyloid beta peptides and microtubule-associated protein tau, respectively. Amyloid beta accumulation and abnormal tau tangles are the pathological hallmarks of Alzheimer's disease.32 Mutations of MAPT have also been discovered as one of the main genetic causes of frontotemporal dementia.33 SNCA gene, encoding alpha-synuclein (alphaSyn), was the first gene identified to cause Parkinson's disease.34 We observed that COVID-19 significantly reduced the expression of APP, MAPT, and SNCA in patients without dementia, and their expression levels were associated with that of 20 hub genes. In patients with dementia, however, COVID-19 increased dementia-related gene expression and was positively correlated with the levels of 20 hub genes. These findings suggest that the dementia status before infection may ultimately determine the effects of COVID-19 on cognitive function.
Accumulating evidence suggests that neuroinflammation is a key player in the pathogenesis of Alzheimer's disease.35 The overactive immune response and cytokine storm observed in the acute stage of SARS-CoV-2 infection are believed to contribute to neuronal damage and synaptic dysfunction.36-38 Notably, it was recently reported that coadministration with anti-inflammatory agents, such as steroid, colchicine, and vitamin C, can reduce the incidence and improve the prognosis of COVID-19.39-41 These findings indicate that COVID-19-induced neuroinflammation might exacerbate the evolution of neurodegeneration.
In this study, we observed that SARS-CoV-2 infection increased the infiltration levels of myeloid dendritic cell activated, endothelial cell common myeloid progenitor, macrophage M1, monocyte and T cell CD4 + Th2, and decreased the infiltration level of common myeloid progenitor endothelial cells in patients without dementia. In addition, the Th1/Th2 ratio was strikingly reduced following COVID-19. These results suggested that COVID-19 suppressed Th1 cell-mediated cellular immunity in nondemented patients. Meanwhile, GSEA results further revealed that the signaling pathways associated with complement, Toll-like receptor, and cytokine receptor were enhanced after COVID-19, suggesting that Th2 cell-mediated humoral immunity was activated. Our findings imply that in COVID-19, the immune system of nondemented patients could not produce sufficient cytotoxic T cells and Th1 type cytokines to inhibit virus replication and remove the virus from the host, which results in chronic SARS-CoV-2 infection and immune escape. This compromise between the host and virus can reduce the damage that cytokine storms inflict upon brain nerves. In contrast, an opposite result was observed in COVID-19 patients with dementia. In this situation, SARS-CoV-2 infection promoted a Th1 cell-mediated cellular response and generated large amounts of Th1-type cytokines. This reaction was helpful to clear SARS-CoV-2, but overactive immune responses may further aggravate neuronal damage and cognitive impairment in patients with dementia. This difference in the immune response between dementia and non-dementia may partially explain the distinct effects of COVID-19 on cognition. Moreover, these findings suggest that appropriate anti-inflammatory drugs should be coadministered for COVID-19 patients with dementia, but not recommended for COVID-19 patients without dementia.
Finally, we explored the possible mechanisms responsible for the distinct immune response. GO biological processes analysis indicated enrichment in the MAPK cascade, endothelial cell migration, cellular response to hypoxia, inflammatory response, and lymphocyte-mediated immunity. MAPK signaling pathway is a highly conserved signal transduction pathway that has a major role in regulating innate immunity during pathogen infection.42 Recently, MAPK hyper-activation has been observed in the lung, heart, and platelets of COVID-19 patients.43-45 Hypoxia is a major pathological feature of COVID-19. Studies have indicated that hypoxia can upregulate the expression of COVID-19 receptor ACE2 and increase the production of pro-inflammatory cytokines.46-48 In addition, our data suggest a potential role for peroxisome and autophagy during COVID-19, which are important regulators of amyloid beta clearance and tau hyperphosphorylation.49, 50
There were several limitations to this study. First, although the participants in this study contained COVID-19 patients with and without dementia as well as age-matched controls, the sample size is still small. Second, our findings were based on the whole-transcriptome sequencing data, direct evidence from histopathological changes, such as senile plaques, neurofibrillary tangles, and dementia severity, are lacking. Third, the human brain tissue samples were obtained through autopsy and long-term clinical follow-up data are not available, especially the cognition alteration of patients after SARS-CoV-2 infection. In the future, much needs to be done to authenticate these findings by longitudinal follow-up from a large sample size. It is also important to obtain evidence from cell and animal experiments when the experimental conditions permit.
In summary, the present study uncovered a distinct influence of COVID-19 on cognitive function and immune response between demented and nondemented patients. Furthermore, our findings identified many potential biomarkers and molecular mechanisms responsible for the impacts of COVID-19 on dementia. Overall, our work may serve as an important clinical reference for COVID-19 treatment in elderly patients.
AUTHOR CONTRIBUTIONS
Xiaojiang Hao and Hui Song conceptualized the study. Jue Yang and Hui Song contributed to data collection, performed analysis and interpretation, and wrote the paper draft. Xiaojiang Hao revised the manuscript. All authors contributed to the article and approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (82160813, 32060210, U1812403), the State Key Laboratory of Drug Research (SIMM2105KF-15), Guizhou Provincial Natural Science and Technology Projects (QKHZK[2021]526, QKHZK[2021]448), the Project of Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education, Guizhou Medical University (QJHKY[2020]251) and the Cultivation Project of National Natural Science Foundation of China of Guizhou Medical University (20NSP064 and 19NSP008), (Qian Ke He Ping Tai Ren Cai (2019])5106 and TianchanziJzi (2021)07.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




