The whole-genome sequencing of prevalent DENV-1 strains during the largest dengue virus outbreak in Xishuangbanna Dai autonomous prefecture in 2019
Daiying Li, MingWang Long, and Tingting Li are co-first authors and contributed equally to this study.
Abstract
In 2019, a serious dengue virus (DENV) infection broke out in the Xishuangbanna Dai Autonomous Prefecture, China. Therefore, we conducted a molecular epidemiological analysis in people that contracted DENV serotype 1 (DENV-1) during this year. We analyzed the molecular epidemiology of six DENV-1 epidemic strains in 2019 by full-length genome sequencing, amino acid mutation site analysis, evolutionary tree analysis, and recombination site comparison analysis. Through the analysis of amino acid mutation sites, it was found that DENV-1 strain (MW386867) was different from the other five epidemic DENV-1 strains in Xishuangbanna in 2019. MW386867 had unique mutation sites at six loci. The six epidemic DENV-1 strains in Xishuangbanna in 2019 were divided into two clusters. MW386867 was highly similar to the MG679800 (Myanmar 2017), MG679801 (Myanmar 2017), and KC172834 (Laos 2008), and the other five strains were highly similar to JQ045660 (Vietnam 2011), FJ176780 (GuangDong 2006). Genetic recombination analysis revealed that there was no recombination signal in the six epidemic DENV-1 strains in Xishuangbanna in 2019. We speculate that the DENV-1 epidemic in 2019 has a co-epidemic of local strains and cross-border strains.
1 INTRODUCTION
Dengue virus (DENV) is a member of Flavivirus of Flaviviridae, which is a type of arboviruses transmitted mainly by Aedes, especially Aedes albopictus and Aedes aegypti.1 The high incidence of DENV was mainly found in tropical and subtropical areas.1 DENV is a kind of enveloped, single positive chain (+ssRNA) RNA virus with a total genome length of ∼10.7 kb.2 The DENV genome encodes a protein of about 3500 amino acids, which is cleaved by the host protein to form three structural proteins and seven nonstructural proteins.2 The former mainly includes envelope protein (E protein), capsid protein (C protein), and premembrane protein (PrM protein), in which PrM protein is cleaved by the host protease furin to yield mature virions.2 The latter mainly includes nonstructural protein 1 (NS1), nonstructural protein 2A (NS2A), nonstructural protein 2B (NS2B), nonstructural protein 3 (NS3), nonstructural protein 4A (NS4A), nonstructural protein 4B (NS4B), and nonstructural protein 5 (NS5).2 There are four antigenically different serotypes of DENV based on the differences in their viral E protein: DENV serotype 1 (DENV-1), DENV-2, DENV-3, and DENV-4.1, 2 In recent years, with the warming of the global climate and the development of transportation systems, the epidemic areas infected with DENV have gradually expanded worldwide, and the rates of infection and mortality have increased, mainly concentrated in some countries in Southeast Asia, the Americas, the Western Pacific,1, 3 Xishuangbanna Dai Autonomous Prefecture (XDAP) in Yunnan province (China), and Guangdong province (China).4 In 1978, the first case of DENV infection was detected in Guangdong province.4 In the following years, several cases of DENV infection were detected in Zhejiang, Fujian, Guangdong, Yunnan, and Guangxi provinces in China.4 In 2018, some Southeast Asian countries adjacent to China successively reported cases of DNV infection, including Vietnam, Laos, Cambodia, Myanmar, Thailand, and Malaysia.4 XDAP is a tourist city in the Yunnan province, bordering Myanmar and Laos. Due to its unique climate and geographical location, numerous tourists come to travel every year. Meanwhile, the climate is very suitable for the growth and reproduction of Aedes mosquitoes, so it has become one of the main areas where dengue fever is prevalent. From 2013 to 2017, new cases and small-scale regional outbreaks occurred almost every year in XDAP.5-8 In 2013, 2015, and 2017, 1262,9 1131,5 and 118410 confirmed DENV cases were reported, respectively. A DENV-1 epidemic occurred from October to December 2018 in XDAP.11 Due to its proximity to the border, unique climate, various insect vectors, and complex cross-border population, the number of imported cases has increased year by year,5, 9, 10 posing a great challenge to the prevention and monitoring of DNV in China and other countries. Globally, about 128 countries have reported cases of DENV infection, covering 390 million people, of whom 96 million people showed different clinical symptoms.12-14 About 500 000 people annually require hospitalization, with a fatality rate of about 2.5%,13 which has imposed great economic pressure on the world's development. Most DENV infections are asymptomatic or mild. Only about 25% of patients with DENV infection had self-limiting symptoms (e.g., fever) and slight blood biochemical abnormalities.14 Only a small number of infected patients will develop into severe symptoms, such as dengue hemorrhagic fever, vascular leakage syndrome, and shock syndrome.14 Severe patients typically appear after re-infection or multiple infections. Because the primary infection can induce the immune response of the body and produce high levels of homotype-specific neutralizing antibodies, it can be effectively protected against the same type and/or heterogeneous DENV infection.2 However, due to the decrease of antibody level, it may promote homotypic or heterosexual virus infection at the subneutralization antibody titer level, resulting in an increase in the number of clinically severe patients.2 This phenomenon is called antibody-dependent enhancement of virus infection.14 Moreover, antibody-dependent enhancement is also the main obstacle to developing effective vaccines for DENV infection.14 At present, due to the lack of specific vaccines and therapeutic drugs in the market, investigation of the origin, molecular mechanisms, and evolution of DENV in the outbreak area are of great significance to prevent DENV outbreaks in the future. In this study, we analyzed the genomic characteristics of the dengue-1 epidemic strain in the large-scale dengue outbreak in Xishuangbanna in 2019, with a view to providing a reference for dengue prevention and monitoring in China and other countries (Table 1).
| Forward primer | Reverse primer | Tm | Amplification length | |
|---|---|---|---|---|
| 1 | CGACCGTCTTTCAATATGC | GCCTTCAGAGGACATCCA | 50 | 1–660 |
| 2 | TTCCCAAACTGGCGAGCA | GGTGTGAACGGTGACTATTACT | 55 | 582–1275 |
| 3 | CGTGTGCTAAGTTCAAGTGTG | GGTGCATCTGTTCCTTCGTAT | 54 | 1184–1835 |
| 4 | GCACAGGCTCATTCAAACTAGA | CCGATGGCTGCTGATAGTCT | 55 | 1745–2465 |
| 5 | CACACTTGGACAGAGCAATACA | CTGGTCTGTAATTGTGCTGAGA | 55 | 2401–3100 |
| 6 | CACACTCTATGGAGCAATGGA | CACAGATGCCACTAGACTCAAT | 54 | 3010–3723 |
| 7 | GAGACCGATGTTTGCTGTAGG | AGGAGAATGGTTAGCGTGTCA | 55 | 3630–4331 |
| 8 | GGAAGAAGAAGCAGAACACTCT | GGTTCCACTTGTTGTCACTACT | 54 | 4218–4905 |
| 9 | CCTGAAGGCGAAGTTGGAG | ATCTTGGATAACTGCGTTGCT | 54 | 4874–5524 |
| 10 | AGAGCAACGCAGTTATCCAAG | AGCACCATCTTCTGTCAGAGTA | 55 | 5408–6118 |
| 11 | TCTACCTGTCTGGCTATCCTAC | CATCTCATTGGCTGCTACTGT | 54 | 6051–6741 |
| 12 | TTCCAGAGCCAGACAGACAA | CGTGATAGACTCGCACAAGG | 54 | 6641–7320 |
| 13 | GGACCCTGTGGTTTATGATGC | GGAGAGGACTCACCAATATCAC | 54 | 7200–7934 |
| 14 | TGGCGACCTATGGATGGAAT | GTGTAGTGTCAGTCATGGCTAT | 54 | 7826–8518 |
| 15 | GGTCAATGGCGTGGTGAA | GGTCATCCTCTGTTATCCTTGT | 53 | 8436–9115 |
| 16 | GAGAAGGACTCCACAAACTTG | CTCCATCCAGCACCTTGT | 52 | 9002–9716 |
| 17 | TGACTGCTCTGAATGATATGGG | GCCTGACTTCTTGCCTTGTT | 54 | 9603–10 331 |
| 18 | TCAGTGTGGAATAGGGTTTGGA | TGCCTGGAATGATGCTGTAGA | 55 | 10 001–10 735 |
- Abbreviation: DENV-1, DENV serotype 1.
2 MATERIALS AND METHODS
2.1 Collection of samples
During the DENV outbreak in the XDAP in 2019, serum samples were obtained from patients who were admitted to XDAP People's Hospital from September to November 2019 due to DENV-NS1. We finally harvested 3900 serum samples (DENV-NS1-positive) in total.
2.2 Whole-genome amplification and sequencing
In the present study, 5 ml of blood samples were obtained from DENV-NS1-positive patients. Then, serum was harvested after centrifugation at 4000 rpm for 15 min, and preserved at −80°C. Viral RNAs were extracted from serum using a QIAamp Viral RNA Mini kit. A PrimeScript™ RT Reagent kit (RR047A) was used to perform reverse transcription to obtain cDNA. The whole-genome sequencing was carried out by designing primers according to sequences of DENV-1 reference strains (MN923084 and MF681693). All primers were synthesized and purified by TsingKe Biological Technology Co., Ltd. The primer sequences are listed in Section 3.3.
Six complete genome sequences of epidemic DENV-1 strains were successfully obtained from human samples and were uploaded into the NCBI GenBank database (Accession Nos. MW386862, MW386863, MW386864, MW386865, MW386866, and MW386867). These DENV sequences were aligned by MEGA 11 using the ClustalW algorithm. A maximum likelihood tree was then constructed using MEGA 11, with 1000 bootstraps. We constructed the distinct phylogenetic trees using six complete genome sequences of epidemic DENV-1 strains in Xishuangbanna in 2019 and 48 reference DENV-1 complete genome sequences (Supporting Information: Table 1).
2.3 Analysis of molecular characteristics and protein secondary structure
The difference in the secondary structure between the structural proteins of the standard DENV-1 strain was calculated using the PredictProtein database (website: https://www.predictprotein.org/). The amino acid sequences and potential RNA, DNA, nucleotide, and protein binding sites were then analyzed.
2.4 Recombination analysis
The analysis of recombination and molecular evolution was conducted using the RDP5.05 package.15 In the recombination analysis, the reference viral sequences were derived from the GenBank sequence database according to geographically close viral sequences or phylogenetic trees using accession numbers below: KR296743, MF681692, MF683116, MF681693, MG679800, MG679801, KY937185, KY937187, KY937188, KY937189, HQ624984, KC172829, KC172835, KC172834.1, JQ045660, FJ390379.
3 RESULTS
3.1 Retrospective summary and analysis of DENV infection in Xishuangbanna from 2013 to 2019
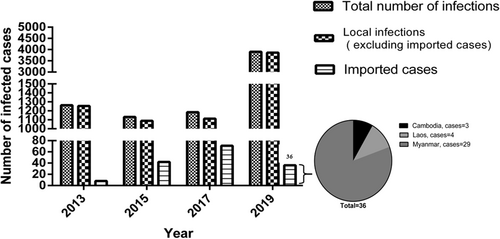
Since the establishment of the DENV infection monitoring system in XDAP in 2008, uninterrupted cases of DENV infection have been annually monitored. The first outbreak occurred in 2013 and the largest outbreak occurred in 2019. It is noteworthy that DENV infection showed a trend of outbreak every other year. Therefore, based on previous studies, we found 1262 infected cases (mainly DENV-1 and DENV-3 infections) in 20139 (Figure 1), of whom there were 8 cross-border imported cases. In 20155 (Figure 1), there were 1131 cases of infection (mainly DENV-2 infection), including 42 cross-border imported cases. In 201710 (Figure 1), there were 1184 infected cases (mainly DENV-1 infection), involving 71 cross-border imported cases. In 201911 (Figure 1), 3900 cases of infection (mainly DENV-1, DENV-2, and DENV-3 infections) were detected, of whom 36 cases were admitted to XDAP People's Hospital from September to November 2019. Then, the sources of cross-border imported cases among infected patients in 2019 were statistically analyzed, including 3 cases from Cambodia, 4 cases from Laos, and 29 cases from Myanmar (Figure 1).
3.2 Analysis of amino acid mutation sites of epidemic DENV-1 strains in Xishuangbanna in 2019
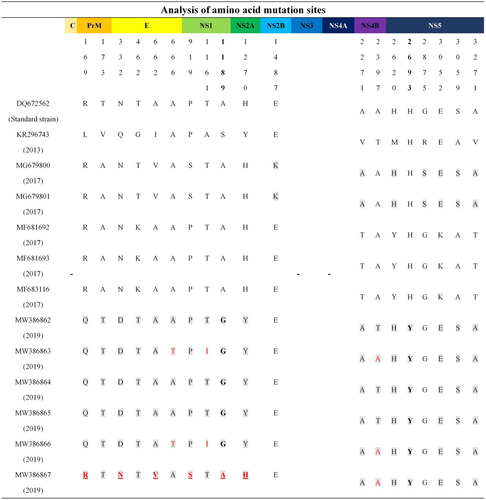
In Xishuangbanna, the largest epidemic DENV infection occurred in 2019, accompanied by the co-epidemic of DENV-1, DENV-2, and DENV-3.11 To investigate the variations of DENV-1 amino acid sites, the amino acid sequences of six epidemic DENV-1 strains isolated in 2019 were compared with the standard DENV-1 strain (DQ672562), and epidemic DENV-1 strains prevalent in Xishuangbanna in 2013 (KR296743) and 2017 (MG679800, MG679801, MF681692, MF681693, and MF683116). The same-sense mutation sites were observed in C, NS3, and NS4A proteins (Figure 2). However, there were different degrees of missense mutation sites in PrM, E, NS1, NS2A, NS2B, NS4B, and NS5 proteins (Figure 2). The mutations in NS2B (1487 amino acid site) and NS5 (2875 amino acid site) proteins were consistent with the amino acid sequences of the standard DENV-1 strain, the DENV-1 prevalent strain in 2013, and the prevalent strains of DENV-1 in 2017 (MF681692, MF681693, and MF683116), which could be reversion mutations (Figure 2). There is no missense mutation among the six DENV-1 strains popular in 2019 in the PrM (173 amino acid site), E (462 amino acid site), NS4B (2271 amino acid site), and NS5 (2620, 2693, 3052, 3059, 3271 amino acid sites) proteins. However, the NS5 (2693 amino acid site) of the six epidemic DENV-1 strains in 2019 was mutated from H to Y, which is different from the epidemic DENV-1 strains in 2013, 2017, and the standard DENV-1 strain. It could be a new missense mutation site. In addition, it was found that MW386867 in 2019 was different from the other five epidemic DENV-1 strains (Figure 2). MW386867 had six unique sense mutation sites, including a PrM protein 169 amino acid site change from Q to R. The 332 and 662 amino acid sites of the E protein changed from D to N and from A to V, respectively. The 919 and 1189 amino acid sites of NS1 protein changed from P to S and from G to A, respectively. The 1270 amino acid site of NS2A protein changed from Y to H. Finally, it was revealed that five DENV-1 strains prevalent in 2019 (MW386862, MW386863, MW386864, MW386865, and MW386866) had a unique amino acid mutation (mutation to G) in NS1 protein (1189 amino acids), which did not appear in the standard DENV-1 strain, or in DENV-1 strains prevalent in 2013 and 2017.
3.3 Phylogenetic analysis of six epidemic DENV-1 strains in Xishuangbanna in 2019
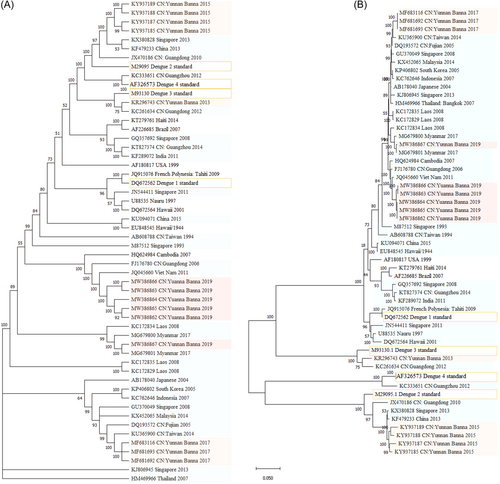
According to the results of phylogenetic analysis, the six epidemic DENV-1 strains in Xishuangbanna in 2019 were divided into two groups. The results of the Bootstrap tree (Figure 3A) and Original tree (Figure 3B) were consistent, representing the distance of kinship and genetic distance, respectively. MW386862, MW386863, MW386864, MW386865, and MW386866 were grouped into a cluster, which was found highly similar to JQ045660 (Vietnam, 2011) and FJ176780 (GuangDong, China 2006) (Figure 3A,B). MW386867 was classified into another cluster, which was found highly similar to MG679800 (Myanmar, 2017), MG679801 (Myanmar, 2017), and KC172834 (Laos, 2008) (Figure 3A,B). The results of the phylogenetic analysis (Figure 3A,B) were consistent with those of amino acid mutation site analysis (Figure 2), which confirmed that MW386867 had a low similarity with the other five strains. To sum up, DENV-1 might have multiple epidemic strains in Xishuangbanna in 2019.
3.4 Recombination analysis of six epidemic DENV-1 strains in Xishuangbanna in 2019
Based on the analysis of the phylogenetic and amino acid mutation sites, we attempted to determine whether the structural and nonstructural proteins of six epidemic DENV-1 strains in Xishuangbanna in 2019 could have a homologous recombination. Therefore, we compared the six epidemic DENV-1 strains in Xishuangbanna in 2019 with the KR296743, MF681692, MF683116, MF681693, MG679800, MG679801, KY937185, KY937187, KY937188, KY937189, HQ624984, KC172829, KC172835, KC172834, JQ045660, FJ390379 using SimplePlot (Supporting Information: 2) and RDP4 (Figure 4), while no significant homologous recombination signals were found. Although MW386865 had weak recombination signals (Figure 4), it still did not meet the requirements of homologous recombination analysis. It was speculated that there was no significant evidence of homologous recombination among the six epidemic DENV-1 strains in Xishuangbanna in 2019.

3.5 Possible secondary structures in structural protein regions
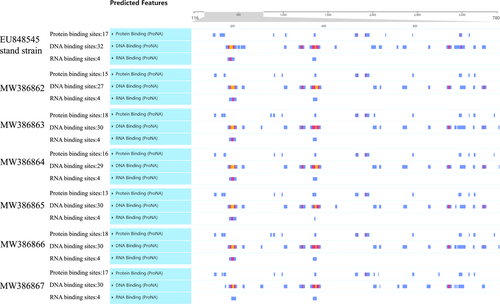
C/PrM/E (structural protein) of DENV is related to virus infection, replication, proliferation, and pathogenicity. Based on the analysis of amino acid mutation sites of the virus, it was found that the C protein of the virus was relatively stable without mutation sites (Figure 2). Therefore, we analyzed the secondary structures of structural proteins (PrM/E). The secondary structures of MW386862, MW386863, MW386864, MW386865, MW386866, and MW386867 were predicted, and 46, 52, 49, 47, 52, and 49 binding sites were found, respectively (Figure 5). In addition, MW386863, MW386864, and MW386866 had unique protein binding sites on 775aa (Figure 5). MW386865 was consistent with the standard strain and had a unique DNA binding site on 425aa from 428aa, while the other five strains did not have this site (Figure 5). The RNA binding site was relatively stable, which was consistent with five epidemic DENV-1 strains and standard DENV-1 strains except for MW386867. MW386867 has only two RNA binding sites. The comparison of the RNA binding sites of MW386867 with that of the standard DENV strain revealed that MW386867 had only a unique DNA binding site on 196aa from 205aa (located in PrM protein) and on 376aa from 383aa (located in E protein) (Figure 5), which could be related to the unique mutation sites of PrM and E protein amino acids (Figure 2).
4 DISCUSSION
XDAP is a tourist city at the southern end of Yunnan province, bordering Myanmar in the West and Laos in the south. Because of its unique climate and geographical location, Xishuangbanna's tourism industry is particularly developed, and millions of tourists annually visit this city. Due to the great number of tourists and the unique tropical monsoon climate, this area has become one of the main areas of epidemic DENV infection. Therefore, for the epidemiological investigation of DENV, preventive and control measures are particularly important. We found that the number of cross-border imported cases and the total number of infected cases have annually increased since 2013 (Figure 1). In 2019, the largest outbreak of infection in history occurred in XADP. In our previous published studies (246 cases),11 we found that the proportion of DENV-1, DENV-2, DENV-3, and DENV-4 was 67.5% (166 cases), 32.1% (79 cases), 0.4% (1 case), and 0% (0 cases), respectively. The ratio of DENV-1:DENV-2 is about 2:1, and there is a co-epidemic dominated by DENV-1 and DENV-2. Therefore, the full-length genome of the six epidemic DENV-1 strains in XDAP in 2019 was amplified, and the evolutionary tree of DENV-1 strains in 2013 and 2017 in the Southeast Asian countries (such as Laos, Vietnam, Myanmar), Yunnan province, and other regions was analyzed (Figure 2). It was found that the six epidemic DENV-1 strains in Xishuangbanna in 2019 were divided into two clusters, in which MW386867 was highly similar to the MG679800, MG679801, and KC172834, and the other five strains were highly similar to JQ045660 (Vietnam) and FJ176780 (GuangDong, China) (Figure 3). To sum up, there might be multiple epidemic DENV-1 strains in Xishuangbanna in 2019, including the local epidemic strain and cross-border imported strains. This result is similar to our previous research,11 that the DENV outbreak in XDAP in 2019 may be caused by the co-circulation of various types of DENV, the increase in the import of cross-border cases, and the co-circulation of cross-border and local strains. Therefore, it is essential to strengthen the participation of different populations in actions to prevent and control mosquitoes, improve the detection of cross-border personnel externally, and strictly prevent the import of cross-border strains. After comparing the amino acid sequences of the six epidemic DENV-1 strains in 2019 with standard DENV-1 strain (DQ672562), epidemic DENV-1 strains in Xishuangbanna in 2013 (KR296743) and 2017 (MG679800, MG679801, MF681692, MF681693, and MF683116), same-sense mutation sites were found in C, NS3, and NS4A proteins. However, there were different degrees of missense mutation sites in PrM, E, NS1, NS2A, NS2B, NS4B, and NS5 proteins. The amino acid sequence and nucleic acid sequence of the PrM protein are highly conserved, which can also induce the body to produce antibodies with poor neutralization and an enhanced infection activity, resulting in the viral escape.16-18 E protein can be divided into three domains, which can mediate the viral infection process and include a neutralizing epitope.19 We found that MW386867 popular in 2019 was different from the other five DENV-1 popular strains. MW386867 has a unique meaningful mutation site (Figure 2) at six loci. For example, the 169 amino acid site of PrM protein changed from Q to R. The 332 and 662 amino acid sites of the E protein changed from D to N and from A to V, respectively. The 919 and 1189 amino acid sites of NS1 changed from P to S and from G to A, respectively. The 1270 amino acid site of NS2A changed from Y to H. Nonstructural proteins may be related to the replication and pathogenicity of DENV. For example, NS1 is related to intracellular signal transduction, and NS4B may be associated with the inhibition of the interferon signaling pathway.20 We, in the present study, found that five epidemic DENV-1 strains in 2019 (MW386862, MW386863, MW386864, MW386865, and MW386866) had a unique amino acid mutation (mutation to G) in NS1 (1189aa), which was not found in standard DENV-1 strain (DQ672562), as well as in epidemic DENV-1 strains in 2013 (KR296743) and in 2017 (MF681692, MF681693, and MF683116) (Figure 2). Several previous studies have confirmed that different site mutations in flaviviruses could enhance pathogenicity to a certain extent.21-24 Genetic recombination is one of the important mechanisms for viral evolution to obtain gene mutations. The probability of genetic recombination is noticeably greater than that of natural selection alone and mutation.25, 26 However, through the recombination analysis of the six epidemic DENV-1 strains in 2019, it was revealed that although MW386865 had weak recombination signals, it still did not meet the requirements of the homologous recombination analysis (Figure 4). Therefore, it was speculated that there was no significant evidence of homologous recombination among the six epidemic DENV-1 strains in Xishuangbanna in 2019. Although the scope of DENV infection has spread rapidly worldwide in recent years, causing millions of people to be infected every year, there is still a lack of an approved DENV vaccine or specific antiviral drugs and treatment methods to control DENV fever. Therefore, it is particularly important to investigate the molecular epidemiological characteristics of the DENV. Due to the unique geographical conditions, there will be a great number of infected cases every year in Xishuangbanna. The present study provided a reliable reference for the prevention and control of DENV, and it is essential to further study the molecular mechanism and pathogenicity of DENV, as well as the development of effective vaccines for DENV.
AUTHOR CONTRIBUTIONS
Daiying Li, MingWang Long, and Qiangming Sun designed the research. Yun Shu, Xiyun Shan, Dehong Ma, Tingting Li, and Pinghua Liu collected blood samples. Yue Pan and Junying Chen performed a literature review. Fan Jia, Xiaodan Wang, Juan Zhang, and Shuying Long provided the study materials. Daiying Li performed the experiment. Daiying Li and MingWang Long wrote the manuscript and analyzed the data. Qiangming Sun performed the article revision. All authors reviewed and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We would like to thank the staff of the clinical laboratory department of Xishuangbanna Dai Autonomous precision People's Hospital, Xishuangbanna, PR China for their help in collecting blood samples from dengue virus infected patients. We also thank all study participants for their donated blood samples and support. The study was sponsored by the National Natural Science Foundation of China (31970868), the foundation of the CAMS Initiative for Innovative Medicine (2021-I2M-1-036), the Innovation Team Project of Yunnan Science and Technology Department (202105AE160020), Yunling scholar talent project of Yunnan Province (YNWR-YLXZ-2019-008), Yunnan health training project of high-level talents (L-2019030, H-2017052) and Leading talents project of Chuncheng science and technology (2022SCP004).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Six DENV-1 sequences in Xishuangbanna in 2019 were deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/, accession Numbers from MW386862, MW386863, MW386864, MW386865, MW386866, and MW386867).
The protocol was approved by the Ethics Committees of the Institute of Medical Biology, Peking Union Medical College (Beijing, China), and the Chinese Academy of Medical Sciences (Beijing, China). The study was performed as per the 1974 Declaration of Helsinki for Human Research (the date of the last revision is 2000).




