CXCL1 confers a survival advantage in Kaposi's sarcoma-associated herpesvirus-infected human endothelial cells through STAT3 phosphorylation
Abstract
Many cytokines produced by Kaposi's sarcoma-associated herpesvirus (KSHV)-infected cells have been shown to participate in the pathogenesis of KSHV. Determination of the exact role of cytokines in Kaposi's sarcoma (KS) pathogenesis is limited, however, by the difficulty to manipulate the target genes in human endothelial cells. In this study, we sought to elucidate the role of cytokines in KSHV-infected human immortalized endothelial cell line (HuARLT cells) by knockout (KO) of the corresponding target genes using the CRISPR/Cas9 system. The cytokine production profile of KSHV-infected HuARLT cells was analyzed using a protein array, and several cytokines were found to be highly upregulated following KSHV infection. This study focused on CXCL1, which was investigated by knocked out in HuARLT cells. KSHV-infected CXCL1 KO cells underwent increased cell death compared to KSHV-infected wild-type (WT) cells and mock-infected CXCL1 KO cells. Lytic replication was not observed in KSHV-infected WT nor CXCL1 KO cells. Phosphorylation of STAT3 was significantly suppressed in KSHV-infected CXCL1 KO cells. Additionally, inhibitors of STAT3 and CXCL1 induced cell death in KSHV-infected endothelial cells. Our results show that CXCL1 production is required for the survival of KSHV-infected endothelial cells, and the CXCL1 to STAT3 phosphorylation signaling pathway may be a therapeutic target for KS.
1 INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) was discovered in biopsy specimens of human Kaposi's sarcoma (KS).1 KS lesions comprise diverse cell types, and it is thought that spindle cells drive KS pathogenesis.2 Spindle cells are generally considered to be of endothelial origin as they exhibit many endothelial lineage markers.3, 4 Since the proliferation of spindle cells at KS lesions is dependent on the microenvironment, many studies of KSHV have focused on extracellular growth factors and cytokines.5, 6
Difficulties relating to genetic modification of human endothelial cells have stymied elucidation of the exact role of cytokines in KS pathogenesis. In particular, target genes cannot be knocked out in primary human endothelial cells, which are frequently used in KSHV studies. A conditionally immortalized human endothelial cell line (HuARLT cells) was established from human umbilical vein endothelial cells (HUVECs) using two independent immortalizing genes, allowing expansion of human endothelial cells with a primary-like phenotype.7 Previous studies have shown that KSHV-infected HuARLT cells exhibit prominent KS-like features, including spheroid formation and maintenance of KSHV in three-dimensional (3D) culture.8, 9 We hypothesize that knockout (KO) of specific target cytokine genes in HuARLT cells would be a valuable strategy to elucidate the role of specific cytokines in KS pathogenesis. In this study, we found that several cytokines were highly upregulated in HuARLT cells following KSHV infection, and one of these was targeted using the CRISPR/Cas9 system to establish a KO clone in HuARLT cells.
CXCL1, also known as GRO-α, is an 11 kDa small cytokine coded by a gene on chromosome 4. CXCL1 is one of the major chemokines associated with inflammation.10 The primary mechanism of CXCL1 is CXCR2-mediated signaling, which stimulates many downstream cellular targets.11 CXCL1 is produced by endothelial cells, neutrophils, macrophages, epithelial cells, and various tumor cells. While it is primarily thought to function as an inflammatory mediator through chemoattractant action, various other functions have been identified. CXCL1 is associated with diverse tissue fibrosis.12 A previous study of human endothelial cells treated with CXCL1-neutralizing antibodies reported reduced endothelial cell migration, viability/proliferation, and angiogenesis via the ERK pathway.13 Moreover, several studies have suggested that CXCL1 is involved in tumorigenesis.14-17 KSHV encodes a G-protein-coupled receptor (vGPCR) that is activated by two chemokines, namely, IL-8 and CXCL1.18 Since vGPCR is a critical oncogene for the initiation and progression of KS, CXCL1 has only been a stimulator of vGPCR in KSHV pathogenesis. However, vGPCR is one of the proteins expressed during lytic replication of KSHV. Therefore, CXCL1 may function through vGPCR during latent KSHV infection. Although CXCL1 is induced in latent KSHV infection,19 little is presently known about CXCL1 acting independently of vGPCR in KSHV pathogenesis.
Here, we investigated the role of CXCL1 in KSHV-infected human endothelial cells using the established CXCL1 KO HuARLT cell line. KSHV infection increased cell death in CXCL1 KO cells, but not in wild-type (WT) HuARLT cells. Among the CXCL1-related signaling pathways, phosphorylation of STAT3 was significantly suppressed in KSHV-infected CXCL1 KO HuARLT cells compared to that in infected WT cells. Synthetic inhibitors of STAT3 phosphorylation and CXCL1 production selectively induced cell death in KSHV-infected endothelial cells.
2 MATERIALS AND METHODS
2.1 Cell culture and reagents
HuARLT cells7 were cultured in endothelial cell growth medium-2 (EGM-2; PromoCell) with cell growth supplement and 2 µg/ml doxycycline (Sigma-Aldrich). EA.hy926 cells20 were obtained from the ATCC and cultured in high-glucose Dulbecco's modified Eagle's medium (WelGene) supplemented with 10% fetal bovine serum (GenDEPOT) and 1% antibiotics–antimycotic (Thermo Fisher Scientific). All cells were cultured under humidified conditions with 5% CO2 at 37°C. Ciglitazone and Stattic were purchased from R&D Systems and Merck, respectively.
2.2 Virus isolation and infection
KSHV was produced using iSLK BAC16 cells harboring recombinant KSHV BAC16 as described previously.21, 22 HuARLT and EA.hy926 cells were infected with KSHV as previously described.23 KSHV-infected cells were maintained with 50 µg/ml of hygromycin for the persistence of virus infection and were used for experiments 2 weeks after infection. After infection, the expressions of KSHV viral genes were analyzed every 2 weeks. It found that all infected cells expressed LANA under the culture condition with the treatment of hygromycin for up to 3 months, but no lytic gene was expressed.
2.3 Cytokine array
Cells were seeded at 1 × 106 cells in a 100-mm cell culture dish. After 1 day, the culture medium was changed to Opti-MEM™ (Thermo Fisher Scientific). At 48 h after the culture medium change, the supernatant was collected and centrifuged at 2000g for 10 min. Cytokine array analysis was performed using a Proteome Profiler Human XL Cytokine Array Kit (R&D Systems), following the manufacturer's instructions.
2.4 RNA isolation, complementary DNA (cDNA) synthesis, and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA extraction and cDNA synthesis were conducted using a Ribospin II Kit (GeneAll Biotechnology) and PrimeScript™ RT Master MIX (Takara), respectively, according to the manufacturer's instructions. Takara SYBR™ FAST qPCR mix was used for qPCR, which was performed as described previously.24 The following primers were used: CXCL1F, 5′-ATAGCCACACTCAAGAAT-3′ and CXCL1R, 5′-TTGGATTTGTCACTGTTC-3′, for CXCL1; ORF50F, 5′-TGCTGGAGGATGTGTGCATT-3′ and ORF50R, 5′-CTCCGTGAGGATCCGAATAA-3′, for KSHV ORF50; ORF59F, 5′- TTAGCCTGGGAGTCCTTAATC-3′ and ORF59R, 5′- GCACACCTTCCACTTCTA-3′, for KSHV ORF59; ORF26F, 5′-GGAGATTGCCACCGTTTA-3′ and ORF26R, 5′-ACTGCATAATTTGGATGTAGT-3′, for KSHV ORF26; ORF65F, 5′-ACTATCTCGTGTTCTTAATTGC-3′ and ORF65R, 5′-ATGATCCCGCCTTTGAAT-3', for KSHV ORF65; K8.1F, 5′-TAAACCCACAGCCCATAG-3′ and K8.1R, 5′-CCACCACTACAACGACTA-3′, for KSHV K8.1; ORF73F, 5′-AGACACAGGATGGGATGGAGC-3′ and ORF73R, 5′-AGACACAGGATGGGATGGAG-3′, for KSHV ORF73; GAPDHF, 5′-GGTATCGTGGAAGGACTC-3′ and GAPDHR, 5′-GTAGAGGCAGGGATGATG-3′, for GAPDH. All primers were synthesized by GenoTech.
2.5 CXCL1 enzyme-linked immunosorbent assay (ELISA)
Cells were seeded in 6-well culture plates with the appropriate medium. The next day, the culture medium was changed to Opti-MEM™ (Thermo Fisher Scientific). Forty-eight hours after changing the culture medium, the supernatant was collected and CXCL1 was analyzed using a Human CXCL1/GRO alpha DuoSet ELISA Kit (R&D Systems) following the manufacturer's instructions.
2.6 Cell viability and cell death assays
Cell viability was measured by cell counting using the trypan blue exclusion method and WST-1 assay. For cell counting, 2 × 105 cells were seeded in a 6-well culture plate and incubated for 1 day with the appropriate culture medium. For chemical inhibitor treatment, the culture medium was changed to Opti-MEM™ containing the chemical inhibitor or the same volume of vehicle (dimethyl sulfoxide [DMSO]). After 1 day, the adherent cells were harvested and counted using the trypan blue (Sigma-Aldrich) exclusion method. In the WST-1 assay, 1 × 104 cells were seeded in 96-well plates with the appropriate medium. The next day, the medium was changed to Opti-MEM™, including the chemicals inhibitor or vehicle, and 24 h later, cell viability was measured using WST-1 reagents (Sigma-Aldrich) according to the manufacturer's instructions. Cell death was analyzed using an Lactate dehydrogenase (LDH) Cytotoxicity Detection kit (Merck) as described previously with minor modifications.24 To exclude the effect of cell proliferation, the results were normalized based on the optical density of the cell lysates.
2.7 CRISPR/Cas9 system for CXCL1 KO clone
CXCL1-targeting CRISPR RNA (crRNA) (A35533; chr4:73 869 728–73 869 750) and trans-activating CRISPR (tracrRNA) were purchased from Thermo Fisher Scientific. Guide RNA (gRNA) was prepared by annealing crRNA and tracrRNA following the manufacturer's instructions. gRNA and TrueCut™ Cas9 protein (Thermo Fisher Scientific) were transfected into HuARLT cells using Lipofectamine™ CRIPRMAX™ Cas9 transfection reagent (Thermo Fisher Scientific). The gRNA and Cas9 protein were used at a final concentration of 0.15 µM. After 3 days, the transfected cells were harvested, the cell concentration was adjusted to 8 cells per 1 ml, and the cells were seeded into a 96-well plate at 100 µl per well to isolate a single clone.
2.8 PCR interruption analysis for identification of gene mutations
PCR was conducted using primers targeting the CRISPR gRNA-binding site. The gDNA from WT HuARLT and CXCL1 KO cells was isolated using a DNeasy® Blood & Tissue Kit (Qiagen) and adjusted to the same concentration based on NanoDrop™ Lite spectrophotometer (Thermo Fisher Scientific) readings. Primers targeting the gRNA binding site (primer S, 5′-TCAGTGAGTCTCTTCTT-3′; primer AS2, 5′-TTCACACTTTGGATGTTC-3′) and primers including the gRNA binding site (primer S, 5′-TCAGTGAGTCTCTTCTT-3′; primer AS1, 5′-TCATTGGAACGAAGGAAG-3′) were used. PCR was performed using 2× Taq PCR premix (SolGent Co. Ltd.). The amplification cycle was 95°C for 30 s, 45°C for 30 s, 72°C for 60 s, and a final extension at 72°C for 5 min. After amplification, the PCR products were separated on a 2% Agarose-TBE gel.
2.9 Sequencing analysis of the CXCL1 KO through T-vector cloning
For TA cloning, isolated DNA was amplified by PCR using 2× Taq PCR premix (SolGent) with primers that included the KO site (primer F, 5′-TCAGTCAGTGAGTCTCTT-3′; primer R, 5′-GAAGGAAGCAAGGTTACT-3′). T-vector cloning and sequencing analysis were performed as described previously, with minor modifications.22 Homo sapiens chromosome 4, GRch38.p13 primary assembly 73 869 393 –73 871 308, NCBI accession no. NC_000004.12 was used as the reference sequence.
2.10 In vitro tube formation assay
Ten microliters of Matrigel® (BD Bioscience) was added to the wells of a μ-slide angiogenesis plate (ibidi GmbH) and polymerized at 37°C. Then, 1 × 104 HuARLT cells were seeded in wells containing 50 μl of EGM-2 and incubated at 37°C. After 4 h of incubation, random fields were imaged using a Nikon Eclipse TS100 inverted microscope. Three random images were analyzed using WimTube software (Onimagin Technologies SCA).
2.11 Sphere culture of endothelial spheroid formation
Sphere culture was performed following previously reported spheroid culture methods.8, 25 A 1% solution of agarose (Bio-Rad)/phosphate-buffered saline was mixed with the desired medium so that the final agarose concentration was 0.5% and then added to a 96-well plate at 100 μl per well. The agarose was allowed to solidify at room temperature for 30 min. HuARLT cells were then seeded in agarose-coated plates at 4000 cells per well with 100 μl of medium containing 2 μg/ml doxycycline. The plates were then incubated at 37°C for 2–3 days to form spheroids. The spheroids were resuspended in 50 μl of the desired medium containing 2 μg/ml doxycycline, 0.7 mg/ml human fibrinogen (Merck), 0.4% methylcellulose (Sigma-Aldrich), and 0.5 U/ml human plasma thrombin (Merck), and then mixed with 50 μl of Matrigel® (BD Bioscience). The spheroid–fibrin–Matrigel® mixture was then loaded into a 96-well plate. After polymerization, the plate was incubated at 37°C and the medium containing 2 μg/ml doxycycline was added to avoid drying.
2.12 Apoptosis assay by annexin V staining
HuARLT cells were seeded at 2 × 105 cells per well in a 6-well culture plate with EGM-2 and incubated for 1 day. Twenty-four hours after changing the culture medium to Opti-MEM™ containing doxycycline, the cells were harvested, and annexin V staining was performed according to the manufacturer's instructions. The cells were analyzed using a Guava® easyCyte™ flow cytometer (Luminex) and InCyte™ 3.1 software (Luminex)
2.13 Western blot analysis
Western blot analysis was performed as described previously.24 Immunodetection was performed using primary antibodies against HHV8 K8.1, total ERK, total STAT3 (all from Santa Cruz Biotechnology), HHV8 ORF45 (Thermo Fisher Scientific), β-actin (Sigma-Aldrich), total AKT, HHV8 ORF50, total p38 MAPK, phosphorylated p38 (p-38) MAPDK (Thr180/Tyr182) (all from Bioss Inc.), p-AKT, phosphorylated ERK, p-STAT3 (all from Cell Signaling Technology), GAPDH (Cusabio Technology) originating from rabbit, and against HHV8 LNA (Abcam) originating from rat. The secondary antibodies conjugated with horseradish peroxidase were goat anti-mouse IgG, goat anti-rabbit IgG, and goat anti-rat IgG (all from Bethyl Laboratories).
2.14 Immunofluorescence assay (IFA)
IFA was performed as described previously.22 The primary antibodies were HHV8 ORF 57, HHV K8.1 from mouse (Santa Cruz), and HHV8 LANA from rat (Abcam). Alexa Fluor 568-conjugated anti-mouse IgG and Alexa Fluor 568-conjugated anti-rat IgG (Thermo Fisher Scientific) were used as the secondary antibodies.
3 RESULTS
3.1 KSHV-induced expression of CXCL1 in human endothelial cells
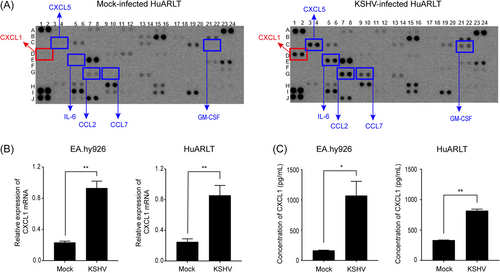
Cytokine production was investigated in mock- and KSHV-infected HuARLT cells using a Proteome Profiler Human XL Cytokine Array Kit. Using this kit, 105 human cytokines in 2-day cultured supernatants from mock- and KSHV-infected cells were analyzed simultaneously. Several cytokines, including CXCL1, ENA-78, IL-6, MCP-1, MCP-3, and GM-CSF, were highly upregulated (Figure 1A). Furthermore, the production of IGFBP-2 (D11 and D12) and GDF-15 (C19 and C20) was suppressed in KSHV-infected cells (Figure 1A). Most of the cytokines that increased in KSHV-infected cells were associated with inflammation, as previously mentioned in other studies of KSHV.26-28 A previous study also showed that KSHV infection of human endothelial cells induces CXCL1.19 However, the exact role of CXCL1 in KSHV pathogenesis still needs to be elucidated. To confirm the increased expression of CXCL1 in KSHV-infected endothelial cells, the messenger RNA (mRNA) expression levels of CXCL1 were analyzed in HuARLT and EA.hy926 cells. Both cell lines exhibited significant enhancement of CXCL1 mRNA expression following KSHV infection (Figure 1B). The protein expression levels of CXCL1 were increased in KSHV-infected HuARLT and EA.hy926 cells (Figure 1C).
3.2 KO of CXCL1 in immortalized human endothelial cells
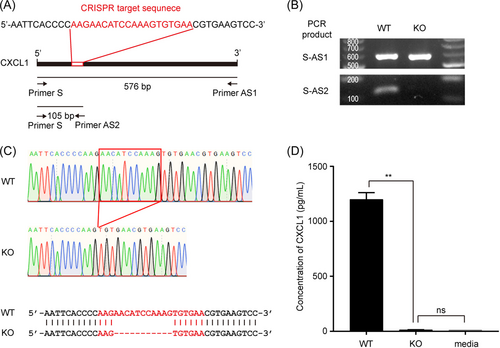
To investigate the role of CXCL1 in KSHV-infected human endothelial cells, we knocked out CXCL1 in HuARLT cells.7 A ribonucleoprotein complex comprising the Cas9 protein and CXCL1-targeting synthetic single-guide RNA was transfected into cells. Each clone was then isolated by limiting dilution, and PCR-based genotyping was used to detect genetic mutations. One pair of primers was designed outside the target sequence (primers S and AS1), whereas the other targeted the Cas9 cut site (AS2). PCR with primer S–AS1 can amplify both WT and mutant alleles (KO), whereas amplification with S–AS2 would be disrupted by new mutations introduced by the CRISPR/Cas9 system. We ran PCR reactions with genomic DNA, designed primer sets from each clone, and chose a clone with disrupted amplification of the S–AS2 PCR products (Figure 2B). To analyze the mutated sequences in this clone, we cloned the PCR products generated using the S–AS1 primers into a T-cloning vector. Ten independent colonies were then isolated and their sequences analyzed using conventional sequencing. We found that all colonies exhibited a deletion mutation at the Cas9 targeting site (Figure 2C). Therefore, we used this clone for further experiments. CXCL1 ELISA indicated that the expression level of CXCL1 in the KO cells was significantly suppressed (Figure 2D). These results indicate that CXCL1 was successfully knocked out in HuARLT cells using the CRISPR/Cas9 system.
3.3 KSHV infection reduced cellular viability in CXCL1 KO HuARLT cells
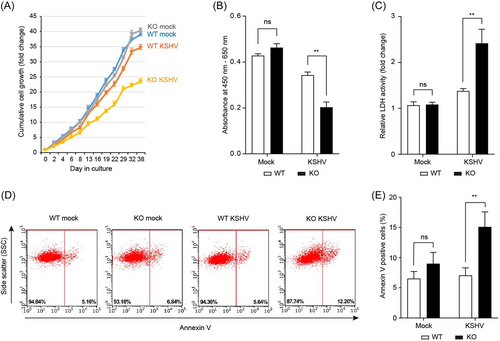
To investigate the effect of CXCL1 on KSHV-infected human endothelial cells, KSHV was infected with WT and CXCL1 KO HuARLT cells. After infection, KSHV-infected cells were selected using hygromycin for at least 2 weeks. The same number of cells were then seeded into a T75 culture flask and subcultured every 2–3 days. Cumulative cell proliferation was analyzed by counting the viable cells in each subculture (Figure 3A). In mock-infected cells, CXCL1 had little effect on cell proliferation. In WT cells, KSHV-infected HuARLT cells exhibited slightly decreased proliferation compared to the mock-infected HuARLT cells. These results may be due to the KSHV infection. However, the possibility that this was the result of the presence or absence of hygromycin B treatment to maintain KSHV infection cannot be excluded. Notably, KSHV infection reduced the proliferation of CXCL1 KO HuARLT cells compared to mock-infected CXCL1 KO cells. The WST-1 cell proliferation assay produced results consistent with the cell counts obtained using the trypan blue exclusion assay (Figure 3B), indicating that viable cell numbers were decreased in KSHV-infected CXCL1 KO HuARLT cells during culture. To determine whether the decrease in the number of viable cells during cell culture resulted from a decrease in cell proliferation or increased cell death, we performed an LDH assay and annexin V staining. The LDH assay demonstrated significantly increased LDH activity in KSHV-infected CXCL1 KO HuARLT cells compared to that in mock-infected KO cells (Figure 3C). More annexin V-positive cells were detected in the KSHV-infected CXCL1 KO HuARLT cell samples than in the mock-infected KO cell samples. Together, these results indicate that KSHV infection-induced cell death in CXCL1 KO human endothelial cells, suggesting that CXCL1 is an essential factor for cell survival in KSHV-infected human endothelial cells.
3.4 CXCL1 KO abrogated KS-related phenotypic features in KSHV-infected HuARLT cells
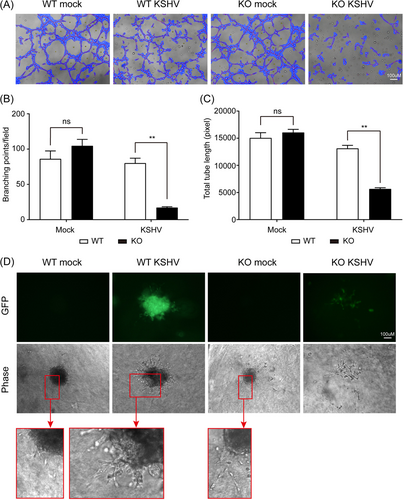
We investigated whether CXCL1 KO affects KS-related phenotypic changes, such as angiogenesis and 3D culture sphere formation. In mock-infected cells, there was no significant difference between WT and CXCL1 KO cells in terms of the number of branch points and total tube length in the in vitro tube formation assay (Figure 4A–C). However, in vitro tube formation was significantly suppressed in KSHV-infected CXCL1 KO HuARLT cells compared to that in KSHV-infected WT cells (Figure 4A–C). These results suggest that CXCL1 is an important factor in angiogenesis during KS development. Previous studies showed that KSHV-infected HuARLT cells could aggregate into spheroids, and the embedded spheroids exhibited pronounced sprouting and invasion of the surrounding matrix in 3D culture.9, 29 Our study also showed that HuARLT cells aggregated into spheroids, and the spheroids from KSHV-infected WT cells sprouted into the surrounding matrix (Figure 4D). Interestingly, while mock-infected CXCL1 KO HuARLT cells aggregated into spheroids and were maintained in a 3D culture matrix, KSHV-infected CXCL1 KO HuARLT cells did not aggregate as well as mock-infected cells. Furthermore, small-sized spheroids of KSHV-infected CXCL1 KO HuARLT cells were scattered in 3D culture within a few days, eventually making them difficult to discern (Figure 4D and Supporting Information: Figure 1). These results suggest that CXCL1 might be critical for KS tumor development.
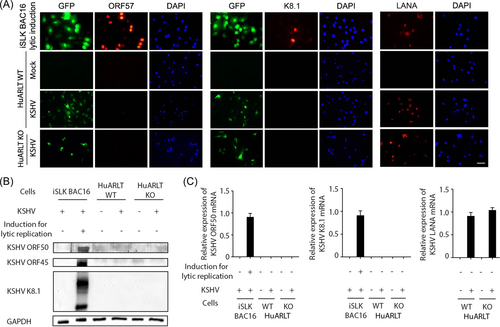
3.5 CXCL1 KO did not induce lytic replication in KSHV-infected HuARLT cells
To determine whether CXCL1 stimulates lytic replication of KSHV, we analyzed viral gene expression in KSHV-infected WT and CXCL1 KO HuARLT cells. Most cells expressed the KSHV latent gene LANA based on IFA (Figure 5A). However, we could not detect the expression of KSHV lytic genes, including KSHV ORF57 and K8.1, in KSHV-infected CXCL1 KO HuARLT cells (Figure 5A). A number of KSHV lytic genes were also analyzed by western blot to confirm viral gene expression. We did not detect KSHV ORF50, KSHV ORF45, or KSHV K8.1 proteins in either KSHV-infected WT or CXCL1 KO HuARLT cells (Figure 5B). There was no KSHV ORF50 or K8.1 mRNA expression in KSHV-infected HuARLT cells, which is consistent with the lack of protein expression (Figure 5C). These results demonstrate that KSHV was maintained in HuARLT cells as a latent infection, and CXCL1 KO did not induce the expression of KSHV lytic genes in HuARLT cells.
3.6 Cell death of KSHV-infected CXCL1 KO HuARLT cells was associated with STAT3 phosphorylation
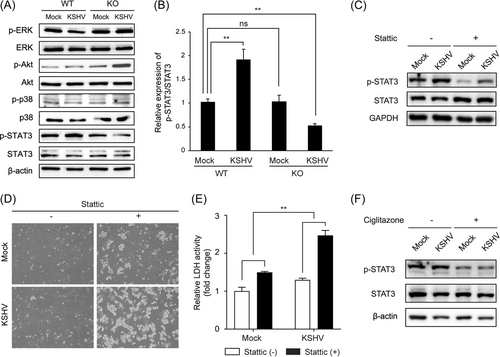
To determine which signaling pathways are associated with cell death in KSHV-infected CXCL1 KO cells, we analyzed the expression of downstream signaling molecules of CXCL1, including ERK, Akt, p38, and STAT3. To exclude the effect of growth factors in the endothelial cell culture medium, protein expression was analyzed 24 h after the medium had been changed to Opti-MEM™. There was no decrease in phosphorylated proteins, except p-STAT3, in KSHV-infected CXCL1 KO cells (Figure 6A). CXCL1 KO did not affect the expression of p-STAT3 in WT cells. Interestingly, KSHV infection induced the expression of p-STAT3 in WT HuARLT cells. However, p-STAT3 was significantly decreased in CXCL1 KO cells following KSHV infection (Figure 6A,B). To investigate whether STAT3 is required for the survival of KSHV-infected cells, the STAT3 inhibitor Stattic was used to treat mock- and KSHV-infected HuARLT cells. Stattic efficiently inhibited STAT3 phosphorylation in both mock- and KSHV-infected HuARLT cells (Figure 6C). A degree of cell death by treatment with the STAT3 inhibitor was observed with mock-infected HuARLT cells. However, with KSHV-infected cells, most of the cells were killed by Stattic treatment (Figure 6D,E). These results suggest that the STAT3 signaling pathway is necessary for the survival of KSHV-infected endothelial cells. To confirm the effect of CXCL1 on STAT3 signaling in KSHV-infected cells, we treated mock- and KSHV-infected HuARLT cells with ciglitazone, which is known to inhibit CXCL1 expression.30 We found that p-STAT3 was reduced by ciglitazone treatment in mock- and KSHV-infected HuARLT cells (Figure 6F). Taken together, our results demonstrate that KSHV-infected HuARLT cells require activation of the STAT3 pathway for cell survival through the expression of CXCL1.
3.7 CXCL1 inhibitor-induced cell death in KSHV-infected endothelial cells
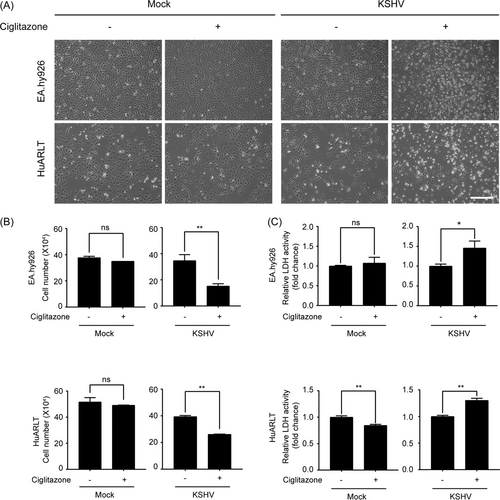
To investigate whether the CXCL1 inhibitor ciglitazone could selectively eliminate KSHV-infected endothelial cells, ciglitazone was used to treat mock- and KSHV-infected endothelial cells. Since EA.hy926 and HuARLT cells exhibited increased expression of CXCL1 following KSHV infection (Figure 1), both cell types were treated with ciglitazone and the effects on the cells were analyzed. Ciglitazone had no adverse effects on the morphology of mock-infected cells after 24 h of treatment. However, many floating cells were observed for KSHV-infected cells treated with ciglitazone (Figure 7A). These results indicate that ciglitazone induced cell death in KSHV-infected cells, but not in mock-infected endothelial cells. To confirm this, the cells were detached using trypsin-EDTA and quantified by trypan blue exclusion assay. The number of viable mock-infected cells was not significantly different between ciglitazone-treated and nontreated cells. However, with KSHV-infected cells, treatment with ciglitazone induced a significant decrease in the number of live cells compared to vehicle (DMSO)-treated cells (Figure 7B). LDH activity also significantly increased only in KSHV-infected cells treated with ciglitazone (Figure 7C). Treatment of ciglitazone or Stattic did not induce lytic replication in KSHV-infected HuARLT cells (Supporting Information: Figure 2). These results indicate that the CXCL1 inhibitor ciglitazone may be a selective death-inducing agent for KSHV-infected endothelial cells.
4 DISCUSSION
Since the discovery of KSHV in 1997, many host genes have been identified as KSHV pathogenesis-associated factors. Many cytokines have been suggested as crucial factors for the microenvironment of KS development. However, to the best of our knowledge, there have been few studies to date using specific target gene–KO human endothelial cells. In a previous study, we knocked out HMGB1 in the KSHV-producing cell line iSLK BAC16 using the CRISPR/Cas9 system, and we elucidated the role of HMGB1 in KSHV production.22 Because the iSLK cell line is a rapidly proliferating cancer cell line, there were no technical impediments to the application of the CRISPR/Cas9 system and isolation of the KO clone. However, the same method for isolating KO clones could not be applied to primary human endothelial cells because replicative senescence is frequently induced during the limiting dilution process. Therefore, we used immortalized human endothelial cells (HuARLT cells) to isolate CXCL1 KO clones. Using the limiting dilution processes, we successfully isolated several CXCL1 KO clones, and the KO was confirmed by sequencing and ELISA for CXCL1. Our results demonstrated that the isolation of a specific target gene KO clone using the CRISPR/Cas9 system was possible with immortalized human endothelial cells, which may provide an opportunity to elucidate the exact role of a host gene in KSHV pathogenesis.
After KSHV infection, we found that the expressions of several cytokines were changed. Among them, we focused on the CXCL1 because it is one of the most highlighted cytokines in constituting the tumor microenvironment in previous studies. CXCL1 from tumor-associated macrophages and cancer-associated fibroblasts enhances tumor growth through the interaction among cancer cells and stromal cells.31 CXCL1 can also increase cell migration and invasion of prostate cancer through AKT/NF-κB signaling.32 We hypothesized that CXCL1 expressed from KSHV-infected cells might play a role in KS development. We showed that CXCL1 plays a vital role in the survival of KSHV-infected cells, but further study is needed to determine what role CXCL1 plays in KS development in the microenvironment of animal models in the future.
To the best of our knowledge, this is the first study to demonstrate that CXCL1 induced by KSHV infection contributes to the survival of latently KSHV-infected human endothelial cells through activation of STAT3 phosphorylation. Previous studies have indicated that KSHV infection induces phosphorylation of STAT3 at serine 727 (S727) and tyrosine 705 (Y705) in human endothelial cells.33, 34 Our results consistently showed increased phosphorylation of STAT3 at Y705 in WT HuARLT cells after infection (Figure 6A–C,F). However, KSHV infection did not induce phosphorylation of STAT3 in CXCL1 KO cells. Since CXCL1 is known to phosphorylate STAT3 at Y705,35 it is not surprising that STAT3 phosphorylation at Y705 is induced in response to CXCL1 upregulation in KSHV-infected endothelial cells. Phosphorylation of STAT3 by CXCL1 appears to be required for KSHV-infected endothelial cells to overcome the stress induced by KSHV infection. While inhibition of STAT3 phosphorylation by Stattic or ciglitazone suppressed p-STAT3 in both mock- and KSHV-infected cells, more cell death was observed in KSHV-infected cells than in mock-infected cells. These results suggest that STAT3 phosphorylation is more critical for the survival of KSHV-infected endothelial cells than in mock-infected cells. STAT3 has known to regulate the KAP1, which represses lytic genes of KSHV.36 These results implicate that STAT3 is an important cellular prosurvival and proproliferative protein constitutively active in KSHV+ tumors. In PEL cells, the inhibition of Y705 constitutive phosphorylation of STAT3 activates the p53–p21 axis and KSHV lytic cycle.37 Considering these previous results, it seems possible for KSHV-infected CXCL1 KO HuARLT cells to induce lytic replication because CXCL1 KO HuARLT cells with KSHV showed Y705 phosphorylation of STAT3 was suppressed compared to the mock-infected CXCL1 KO cells. However, lytic replication was not induced in CXCL1 KO HuARLT cells (Figures 5 and 6). Our results indicate that inhibiting Y705 phosphorylation of STAT3 alone is insufficient to induce lytic replication in HuARLT cells. In other words, CXCL1-mediated STAT3 phosphorylation in KSHV-infected HuARLT cells would provide a survival signal independent of lytic replication. Nevertheless, based on the previous studies, CXCL1-mediated STAT3 phosphorylation may participate in the maintenance of latent replication of KSHV.
KSHV infection usually increases tube formation in human endothelial cells.38, 39 However, in our experimental conditions, we found similar or less tube formation activity in KSHV-infected HuARLT cells than in mock-infected cells. It is not known exactly why this difference is from the previous studies, but the combination of HuARLT cells and KSHV BAC16 could be the cause. A previous study demonstrated that HuARLT cells are immortalized cell lines with lower tube formation activity than the originated cells, HUVECs.40 In the same study, rKSHV.219 infection overcame the less tube formation activity of HuARLT cells and showed an increase in tube formation in HuARLT cells. Our study used KSHV BAC16 instead of rKSHV.219 for infection. Further experiments are required to determine whether different recombinant KSHV induces differences in angiogenesis-associated gene expression in HuARLT cells. Decreased in vitro tube formation and defective 3D culture of the spheroid of KSHV-infected CXCL1 KO cells would be due to cell death. Because cell death was significantly increased in KSHV-infected CXCL1 KO HuARLT cells, we did not analyze the expression of growth factors or cytokines from these cells. However, we cannot completely rule out the possibility that other factors may be associated with these results.
Several previous studies have shown that CXCL1 is upregulated in KSHV infection.19, 41 We also observed that CXCL1 was highly upregulated in HuARLT and EA.hy926 cells by KSHV infection. However, it is still unknown which viral gene is associated with the induction of CXCL1 expression. Since KSHV-infected HuARLT cells did not show expression of genes related to lytic replication of KSHV, one or a combination of latent KSHV genes may affect CXCL1 expression. Moreover, the mechanism underlying CXCL1 expression by KSHV infection should be investigated in future studies. Despite this limitation, this study demonstrated that CRISPR/Cas9 system-mediated human endothelial cell KO clones may be a valuable tool to study KSHV–host interaction and that the production of CXCL1 is critical for the survival of latently KSHV-infected human endothelial cells and persistent viral infection. Thus, CXCL1 may be a valid target for KS treatment.
AUTHOR CONTRIBUTIONS
Myung-Ju Lee and Myung-Shin Lee conceived the research design. Myung-Ju Lee, Jisu Lee, Su-Kyung Kang, Seung-Min Yoo, and Changhoon Park conducted experiments and acquired data. Dagmar Wirth advised on the study design and in preparing the manuscript. Myung-Ju Lee and Myung-Shin Lee wrote the main manuscript. All authors reviewed the manuscript.
ACKNOWLEDGMENTS
This research was supported by the National Research Foundation of Korea (NRF), project number NRF-2019R1A2C2083947 and NRF-2022R1I1A3065538 (to M.-S.L.).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




