Differential avidity determination of IgG directed towards the receptor-binding domain (RBD) of SARS-CoV-2 wild-type and its variants in one assay: Rational tool for the assessment of protective immunity
Abstract
The avidity (binding strength) of IgG directed towards the receptor-binding domain (RBD) of spike protein has been recognized as a central marker in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serology. It seems to be linked to increased infection-neutralization potential and therefore might indicate protective immunity. Using a prototype line assay based on the established recomLine SARS-CoV-2 assay, supplemented with RBD of the delta and the omicron variant, differential avidity determination of IgG directed towards RBD of wild-type (WT) SARS-CoV-2 and distinct variants was possible within one assay. Our data confirm that natural SARS-CoV-2 infection or one vaccination step lead to low avidity IgG, whereas further vaccination steps gradually increase avidity to high values. High avidity is not reached by infection alone. After infection with WT SARS-CoV-2 or vaccination based on mRNA WT, the avidity of cross-reacting IgG directed towards RBD of the delta variant only showed marginal differences compared to IgG directed towards RBD WT. In contrast, the avidity of IgG cross-reacting with RBD of the omicron variant was always much lower than for IgG RBD WT, except after the third vaccination step. Therefore, parallel avidity testing of RBD WT and omicron seems to be mandatory for a significant assessment of protective immunity towards SARS-CoV-2.
1 INTRODUCTION
The efficiency of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is dependent on the efficiency of binding between the receptor-binding domain (RBD) of the viral spike protein S1 and the cellular angiotensin-converting enzyme 2 (ACE2) receptor. This process is driven by high affinity between RBD and its cellular receptor.1 To interfere with this binding step, protective IgG therefore not only needs to specifically recognize the RBD domain (a feature, i.e., assessed in classical neutralization tests), but also must inherit a sufficiently high binding strength (avidity) towards RBD, thus being able to compete with RBD/ACE2 binding
In general, a gradual increase in the binding strength (avidity) of IgG to target epitopes is triggered after infections, as well as by vaccinations. This complex, stepwise increase in avidity of the IgG response is based on IgG affinity maturation.2-4 Avidity determination can therefore be used to differentiate between acute infections (low avidity) and past infections (high avidity).5-8 It has also been recognized that establishment of high avidity is a prerequisite for protective immunity in many viral systems.9, 10
Avidity maturation after SARS-CoV-2 infection or vaccination follows a pattern that is different from nearly all other viral infections,10 as natural infection with SARS-CoV-2 does not regularly lead to the establishment of a high avidity IgG response.11-18 The limitation of the IgG response to low or intermediate avidity seems to be part of the specific biology of coronaviruses,19 allowing for repeated waves of reinfection.20, 21
“Incomplete” avidity maturation after SARS-CoV-2 infection is most likely due to the lack of sufficient antigen presentation for a sufficiently long time to drive the completion of the avidity maturation process.10 This assumption is based on (i) results in other biological systems, where induced restriction of antigen availability prevented avidity maturation,22, 23 and by the finding that COVID-19 patients with more severe disease, which are therefore exposed to higher viral loads for longer times, frequently show a gradually higher avidity of IgG directed towards SARS-CoV-2 than patients with the milder disease.12, 17, 24, 25 This picture can, however, be drastically changed in cases of most severe COVID-19 that lead to death. In these cases, avidity maturation can be prevented by SARS-CoV-2 infection through destruction of the germinal centers in secondary lymphatic organs.26
In contrast to natural infection with SARS-CoV-2, prime-boosting vaccination allows for the generation of high avidity IgG.10, 12 It therefore might be essential for the establishment of protective immunity.10 The interval between the first and second vaccination step has been shown to modulate the degree of avidity maturation, as an interval of 16 weeks was leading to higher avidity than an interval of 4 weeks.27 A third vaccination step has been recognized as an additional increase in antibody avidity compared to the second vaccination step, rather than simply maintenance of or increase in an IgG concentration.28, 29 Importantly, priming the immune response by natural infection with SARS-CoV-2, followed by one subsequent vaccination step, leads to very high avidity of the IgG response as well. This is not further increased by an additional vaccination step.12, 27, 29
The work by many groups allowed to establish concepts that help to understand prerequisites for optimal protection toward SARS-CoV-2 infection and disease, and to define individual serological constellations that indicate potential failure of protection. Wratil et al. showed that increased infection-neutralization capacity is associated with higher antibody avidity.29 Moura et al. and Ravichandran et al. presented convincing evidence for a correlation between low avidity and severe outcome of COVID-19 and death.17, 30 Tang et al. defined minimal avidity maturation against the SARS-CoV-2 spike protein as one key immunological parameter of intensive care patients that died from COVID-19.31 In line with this finding, stronger avidity maturation was associated with disease resolution. Finally, Manuylov et al. presented data that indicated that the presence of low-avidity IgG to RBD during reinfection is a negative prognostic factor, in which a patient's risk of developing COVID-19 in a severe form is significantly increased.32 Therefore, though it is certainly not possible to exclude future SARS-CoV-2 infection based on the presence of high avidity IgG directed towards RBD, the lack of high avidity IgG despite vaccination might be regarded as an indication of potential risk. This is particularly relevant for highly exposed, as well as for more vulnerable individuals. In these cases, the determination of a low avidity IgG response towards RBD should be taken as an indication to get additional vaccination, followed by qualified testing of IgG quality.
The present SARS-CoV-2 pandemic is characterized by a continuous generation and selection of viral variants and a replacement of variant populations through new variants. As recently reviewed,10 mutations in the RBD region that caused a continuous increase in the affinity/avidity of RBD for ACE2, thus causing a more efficient infection, seem to be one central determining factor in this development. Though the recent dominant variant omicron carries more than 30 mutations in its spike protein, most of them in RBD, its amazing efficiency of transmission does not seem to depend on further enhancement of RBD/ACE2 affinity.10, 33, 34 Rather, the efficiency of the omicron variant seems to be based on its immune escape phenotype.10, 33-36 This phenotype is impressively demonstrated (i) by lack of the majority of monoclonal antibodies that neutralize SARS-CoV-2 WT to neutralize the omicron variant,10, 34, 36 and by the reduced binding of IgG from convalescent sera to the spike protein.33 In conclusion, RBD of the omicron and the WT strain seem to share only a relatively low number of overlapping, protection–relevant epitopes.34 In addition, the epitopes on omicron RBD that are shared with WT RBD, seem to bind specific IgG with lower avidity than the WT RBD.10, 33 In line with these findings, cross-protection between IgG induced by a WT-specific vaccine and the omicron variant requires at least three vaccination steps, whereas the reaction with the homologous proteins can reach high avidity already after two vaccination steps.29, 35-37
The present epidemiological situation characterized by the complexity of the dynamics of variants, in combination with the focus of serological test systems and vaccines on the WT strain of SARS-CoV-2 demands the establishment of rapid and conclusive test systems. These should also allow determining the quality, that is the avidity of IgG directed towards RBD of the various variants.
2 METHODS
2.1 Patients and sera
Sera were collected from healthy blood donors before November 2019 (uninfected control), polymerase chain reaction (PCR)-confirmed COVI-19 outpatients infected with SARS-CoV-2 WT or the omicron variant, and healthy subjects vaccinated with the BioNTech/Pfizer messenger RNA (mRNA) vaccine (COMIRNATY®/BNT162B2) one, two or three times. Demographic data of these cohorts are summarized in Table 1
| Group | n | Gender | Age (years) | Time of collection (days) | |
|---|---|---|---|---|---|
| F (n) | M (n) | ||||
| Healthy blood donors | 59 | No data | 18–65 | Before November 2019 | |
| Outpatients COVID-19 SARS-CoV-2 (WT) | 24 | 14 | 10 | 18–65 | 15–97 |
| Median = 36 | |||||
| Outpatients COVID-19 SARS-CoV-2 (Omicron) | 14 | 7 | 7 | 18-65 | 17–45 |
| Median = 32 | |||||
| Vaccinees (1X) | 15 | 7 | 8 | 18–49 n = 5 | |
| 50–64 n = 4 | 12–34 | ||||
| 65–79 n = 2 | Median = 18 | ||||
| Vaccinees (2X) | 13 | 3 | 10 | 18–49 n = 7 | |
| 50–64 n = 2 | 10–24 | ||||
| 65–79 n = 2 | Median = 17 | ||||
| >80 n = 2 | |||||
| Vaccinees (3X) | 13 | 8 | 5 | 18–49 n = 8 | |
| 50–64 n = 5 | 32 | ||||
- Note: Table 1 summarizes the patients and sera used in this study. 59 anonymized plasma samples from healthy adult blood donors were purchased from the Bavarian Red Cross. They were assayed to determine the specificity of the new prototype assay. Sera from adult outpatients with clinical signs of COVID-19 (such as fever, headache, loss of smell, sore throat, or pneumonia) either showed SARS-CoV-2 WT or SARS-CoV-2 omicron infection, as determined by RT-PCR. Time of collection (days) refers to the onset of disease in COVID-19 patients. In the case of healthy individuals vaccinated for the indicated times with the COMIRNATY®/BNT162B2 vaccine from BioNTech/Pfizer, time of collection refers to the time (days) after the last vaccination.
- Abbreviations: COVID-19, coronavirus disease 2019; RT-PCR, reverse-transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WT, wild-type.
2.2 Collection and processing of sera
Sera from COVID-19 outpatients and vaccinated individuals were collected in the Munich area after a call for voluntary donation of serum samples for serological analysis related to SARS-CoV-2. The samples were drawn by the family doctors. The volunteers gave their written consent for testing. The logistic support of Mikrogen GmbH collected the sera and all necessary information
The anonymized samples were transferred to Research and Development of Mikrogen GmbH for professional testing in the modified recomLine SARS-CoV-2 IgG line assay, including RBD of SARS-CoV-2 WT, delta variant, and omicron variant. Testing included avidity determination, using our standard conditions (7 M urea, 3 min incubation with urea). For of the members of Research and Development and the first author (G.B.), information was restricted to clinical symptoms, and the time between onset of clinical symptoms and extraction of the sera. Sera were stored at –20°C until they were tested in the immunoassays.
2.3 Immunoblot assay
A. Production of nitrocellulose strips: The conditions for production of the CE-marked recomLine SARS-CoV-2 IgG test (product # 7374 of Mikrogen GmbH) were used, as recently described.11 Individual concentrations of purified recombinant SARS-CoV-2 spike protein-1 receptor-binding domain (RBD) of SARS-CoV-2 Wuhan-Hu-1 (termed “wild-type” (WT) within this manuscript), delta variant (B.1.617.2) and omicron variant (B.1.1.529) were applied directly onto nitrocellulose membranes in separate lanes. Recombinant RBD comprised the following amino acids of S1: Wuhan-Hu-1 (WT): amino acids 319-541; delta variant: amino acids 319-537 (mutations: L452R, T478K); omicron variant (amino acids 319-537 (mutations: G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H). All recombinant proteins contained a His-Tag at C-terminus. For additional internal control, the strips also contained separate lanes with NP and Spike protein-1 (S1) from SARS-CoV-2 (Wuhan-Hu-1), as well as trimeric spike protein from the delta and omicron variant. For control of the technical performance, all strips contained adequate reaction controls, IgG conjugate controls, and cutoff controls. In contrast to the originally described lineassay (product # 7374 of Mikrogen GmbH), no antigens of human seasonal coronaviruses were applied on the test used in this study. Production was standardized and the resultant strips were evaluated.
B. Basic procedure of the line immunoassay: The reactivity of 1:100 dilutions of serum antibodies against the recombinant antigens was detected with peroxidase-labeled anti-human IgG antibody and the use of precipitating tetramethylbenzidine. The first incubation of serum and test strips was for 1 h, followed by three washing steps with buffer. The incubation of the strips with peroxidase-labeled anti-human IgG antibody was for 45 min, followed by three washing steps. Treatment with tetramethylbenzidine was for 8 min.
The line immunoassays were carried out in a semi-automatic processor Dynablot (Dynex Technologies GmbH) with manual serum pipetting according to instruction manual provided by Mikrogen GmbH. An Epson J371A scanner (Epson) and recomScan software (Mikrogen GmbH) were used according to the instruction manuals.
C. Avidity determination: Sera were incubated for 1 h with the recomLine SARS-CoV-2 IgG test strips in duplicate. Then both replicates were incubated for 5 min with wash buffer. One assay was further incubated in wash buffer, while the parallel assay replicate was treated with 7 M urea for 3 min. After three additional washing steps, both assay replicates were processed with anti-human IgG antibody labeled with peroxidase and detected as outlined above to describe the line immunoassay procedure. For the determination of the avidity index, the grey intensity area output by recomScan on the urea treated test strip was divided by the grey intensity of the parallel control assay replicate. The avidity index thus indicates the fraction of IgG that has remained bound after urea treatment. This approach follows the classical established procedure for avidity determination.5, 6 Following Nurmi et al.,6 the border between high and low avidity was defined by an avidity index of 0.6, that is, a condition where 40% of bound IgG were released by standardized urea treatment (7 M urea, 3 min incubation). The optimal conditions for standard avidity determination in our test system have been recently determined.11, 12 For further differentiation between different degrees of avidity maturation, the avidity range below an avidity index of 0.6 has been further dissected into the range of low avidity (avidity index below 0.4) and intermediate avidity (avidity index between 0.4 and 0.6), which is followed by high avidity.11, 12
Precision and reproducibility of the recomLineSARS-CoV-2 assay have been previously described.11
The data analysis by the first author (G. B.) was performed on the basis of raw data. The focus of this initial study with the new prototype lineassay was on the IgG response towards RBD of WT SARS-CoV-2 and its variants delta and omicron. Internal controls and technical controls ensured the quality of performance of the assays.
The Yates continuity corrected Chi-square test (two-sided) was used for the statistical determination of significances (p < 0.01 = significant; p < 0.001 = highly significant).
3 RESULTS AND DISCUSSION
We used a new prototype lineassay, based on the established recomLine SARS-CoV-2 (WT) IgG assay,11, 12 supplemented with RBD of the delta and omicron variant of SARS-CoV-2, as described in the Methods section. As this test system provides antigens from WT, delta, and omicron variant of SARS-CoV-2 on the same stripe, it is suitable for parallel determination of the avidity of IgG directed towards the respective antigens. Fifty-nine prepandemic control sera from individuals without SARS-CoV-2 infection caused no positive signals in this assay (data not shown), ensuring the specificity of this new assay system.
To illustrate the feasibility of the test system for the determination of homologous reactions and cross-reactions, the data obtained for six selected sera per group (infection with SARS-CoV-2 wild-type, infections with SARS-CoV-2 omicron, one to three vaccination steps with the BioNTech/Pfizer mRNA vaccine) are presented. Selection was based on increasing homologous IgG responses towards RBD. The goal was to determine the degree of cross-reactions with RBD of the delta and the omicron variant, in the case of infection or vaccination with SARS-CoV-2 WT, as well as towards RBD of WT and the delta variant after infection with the omicron variant. Sera from nonhospitalized COVID-19 patients showed a significant IgG response towards RBD WT, but no cross-reactive response towards RBD omicron in most cases (Figure 1A). Only one serum showed a marginal cross-reaction. The cross-reactive response towards RBD delta was much stronger than that towards RBD omicron, and it was highly variable. After infection with the omicron variant of SARS-CoV-2, relatively low IgG responses towards RBD omicron were seen, with only marginal cross-reaction with RBD WT or RBD delta (Figure 1B). These data confirm that the use of RBD WT or RBD omicron allows to detect of differential and specific IgG responses. They also show that there is nearly now detectable cross-reaction between IgG directed towards RBD WT and RBD omicron, and vice versa after natural infection. The general picture observed after one vaccination with the BioNTech/Pfizer vaccine (Figure 1C) resembled very much the picture seen for infection with SARS-CoV-2 WT (Figure 1A), in line with our previous findings.12 After two vaccination steps (Figure 1D), the concentration of IgG directed towards RBD WT increased strongly compared to the concentrations reached after one vaccination. Importantly, highly variable degrees of a much stronger cross-reaction with RBD omicron were seen after second vaccination, whereas the concentrations of IgG WT and IgG cross-reacting with RBD delta were nearly identical. The major effect of the third vaccination step was the relative increase in the concentration of IgG cross-reactive with RBD omicron (Figure 1E).
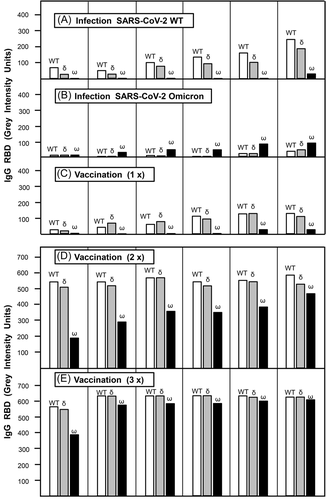
Taken together, these initial data show (i) the power of our test system to differentiate between IgG responses towards RBD WT, delta, and omicron; (ii) the lack of strong cross-reaction between IgG directed towards RBD WT with RBD omicron after natural infection or only one vaccination step, and (iii) the continuous increase in the relative concentration of IgG cross-reactive with RBD omicron after the second and third vaccination step. These features of the test system founded a solid basis for subsequent avidity determination for the evaluation of IgG binding strength, that is, its functional quality. Information on all sera, including data on IgG avidity, from the groups comprising infection with SARS-CoV-2 WT and one to three steps of mRNA-based vaccination with WT can be found in Figures 2 and 3. Data on all sera available after SARS-CoV-2 omicron infection can be found under Supplementary Materials. It will be interesting and necessary to determine the IgG response, particularly its avidity, towards RBD of the omicron and delta variant, as well as of WT after several steps of vaccination with a prospective omicron-based mRNA vaccine. In addition, the outcome of concentration and avidity of IgG directed towards RBD WT, delta, and omicron after heterologous infection/vaccination as well as multiple infections with different strains of SARS-CoV-2 should be in the focus of attention.
Avidity of IgG directed towards RBD is one of the central markers of IgG quality and indicates the potential for protective immunity.10, 17, 29-31 The specific challenge of serology in the case of SARS-CoV-2 infections and vaccinations is due to (i) incomplete avidity maturation after natural SARS-CoV-2 infection, in contrast to the establishment of high avidity after two or more vaccination steps,10, 12 and (ii) the establishment of various variants of the virus. The latter include the immune escape variant omicron which is dominating the pandemics at present. Therefore, qualified serology should allow to determine quantity (IgG concentration) as well as quality (IgG avidity) of homologous as well as heterologous interactions between IgG and target epitopes. As vaccination so far is based on the WT strain of SARS-CoV-2, it is obvious that it is necessary to assess the protective effect of vaccination-induced IgG not only towards WT SARS-CoV-2, but also towards its emerging variants. This aspect is less problematic for variants with a limited number of mutations (like the delta variant) and a resultant less expressed immune escape nature, but particularly complex for the omicron variant due to its strong immune escape nature.
For an analysis of the cross-reactivity and potential cross-protection of IgG towards two selected SARS-CoV-2 types of central significance, we studied the IgG response towards RBD WT and RBD omicron after infection with WT virus, as well as several rounds of vaccination based on a WT virus-based mRNA vaccine. Figure 2A confirms that sera from uninfected individuals gave no signals either with RBD from WT or omicron. The concentration of IgG directed towards RBD WT was of comparable low quantity both after natural infection with WT virus (Figure 2B) or vaccination with a vaccine based on WT (Figure 2C). In both cases, IgG was essentially characterized by low avidity (Figure 2G,H). Though most sera in Figure 2G,H are characterized by an avidity index of zero, Figure 2G shows six sera that are still in the low or intermediate avidity range, but have distinct avidity indices in the range from 0.1 to 0.45, whereas Figure 2H only shows one serum with an avidity index above zero. This difference is due to longer collection times in Figure 2G (median of collection time: 36 days) compared with Figure 2H (median of collection time: 18 days).
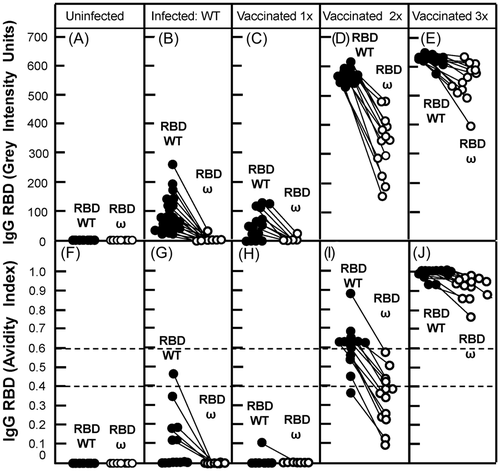
After natural infection or single vaccination there was practically no cross-reaction of IgG directed towards RBD WT with RBD omicron (Figure 2G,H). The second vaccination step caused a steep increase in the concentration of IgG-directed RBD WT, paralleled by a strong increase in its avidity (Figure 2D,I). Thereby 8/13 sera had reached high avidity, 4/13 sera were in the intermediate, and 1/13 in the low avidity range (Figure 2I). In contrast to the situation after the first vaccination step (Figure 2C), cross-reaction with RBD omicron was evident after the second vaccination step (Figure 2D). However, there were significant differences both with respect to the concentration of IgG (Figure 2D) and avidity (Figure 2I) when the reactions towards RBD WT were compared to those towards RBD omicron after second vaccination. None of the cross-reactive sera reached high avidity, whereas 4/13 were in the intermediate and 9/13 in the low avidity range (Figure 2I).
Importantly, the most prominent effect of the third vaccination step was essentially characterized as a further and rather substantial increase in IgG avidities, leading to very high avidity IgG both for the homologous reaction with RBD WT and the cross-reaction with RBD omicron (Figure 2J). This was paralleled by rather high concentrations of IgG reacting with the respective RBDs, as shown by the high grey intensity values (Figure 2E). Therefore, the outstanding benefit from three vaccination steps seems to be the establishment of high avidity IgG that binds to the homologous RBD WT as well as to the heterologous RBD of omicron, as high avidity towards RBD is indicative for the protective function of IgG.17, 29-31 The new prototype recomLineAssay seems to allow a clear dissection between the homologous and the heterologous (cross-reactive) IgG responses, their quantitation, and the measurement of their binding strength (avidity). This should allow to define individuals that did not reach high avidity towards essential SARS-CoV-2 variants despite several vaccination steps. This may be due to trivial problems during vaccination, to the variability of their immune response, or to a major failure of their immune system to promote avidity maturation adequately. Particularly in elderly individuals, deficiencies in the B cell repertoire and delayed avidity maturation might impair the establishment of an adequate response characterized by high avidity.38-40 This rare effect can be predicted to be more severe in the case of cross-reaction with escape variants, as the overlap of epitopes between WT and the variant is limited.34
Infections with either SARS-CoV-2 WT or its variant omicron were leading to reciprocal results for the homologous reactions as well as the respective cross-reactions. After infection with SARS-CoV-2, 23/24 sera showed detectable IgG responses towards WT RBD (Figure S1). 22/23 positive responses were of low avidity, one response was in the intermediary range (Figure S2). Only one serum obtained after infection with SARS-CoV-2 WT showed detectable cross-reaction with RBD of the omicron variant. This cross-reaction was characterized by low avidity (Figures S1 and S2).
After PCR-confirmed infection with SARS-CoV-2 omicron, only 5/14 sera showed detectable IgG directed towards RBD omicron (Figure S1). In all cases, this response was of low avidity (Figure S2). Only two sera showed detectable cross-reaction with RBD of the WT strain, characterized by low avidity (Figures S1 and S2).
These data demonstrate that reciprocal cross-reactions of IgG-directed RBD of SARS-CoV-2 WT and its omicron variant are extremely low after infection with these viruses. This might point to problems for the detection of SARS-CoV-2 omicron infection with test systems based on RBD from WT, and vice versa. The resolution of such diagnostic problems requires the modification and validation of test systems to cope with new and highly mutated variants, as originally suggested by Lippi et al.41
The number of available sera in this pilot study is low, which is one of its weaknesses. However, our data indicate that the percentage of sera positive for IgG directed towards RBD is much lower after infection with omicron compared to the WT. This difference is not explained by different lengths of time after onset of disease, as the medians of the time points of serum acquisition are quite similar (32 vs. 36 days). This aspect deserves attention in subsequent larger studies.
The possibility of our test system to dissect the IgG responses towards SARS-CoV-2 WT and its variants, as shown initially for the omicron variant (Figure 2), raised the question of whether qualified serological testing should include all potentially relevant variants into the test. To address this question, the IgG responses obtained after natural infection or distinct vaccination steps were analyzed with regard to their reaction with RBD derived from WT and the delta variant in the same assay (Figure 3). Measurements were focusing on concentrations of IgG (determined by grey intensity units) (Figure 3A–E) and its quality (determined by avidity measurement) (Figure 3F–J). IgG induced by infection with wild-type SARS-CoV-2, as well as by one to three vaccination steps with mRNA vaccine based on WT virus, reacted with WT RBD and cross-reacted with delta RBD with no or only marginal differences, both with respect to IgG concentration and IgG avidity. This picture was therefore clearly different from the cross-reaction with RBD of the escape variant omicron, as demonstrated in Figure 2. Therefore, in the present pandemic situation, the use of RBD from escape mutants like omicron in addition to WT RBD seems to be mandatory for a significant serological assessment, following the suggestions by Lippi et al.41 However, the parallel use of RBD from variants that only differ from RBD WT in few mutations may give confirmatory evidence, but is neither mandatory nor does it allow to discriminate between infections with SARS-CoV-2 WT and such variants.
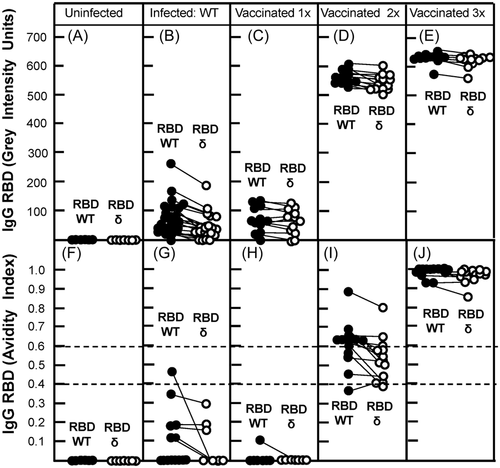
Compared to the SARS-CoV-2 wild-type strain, the Delta variant has been shown to be more transmissible and to have higher viral loads in infected samples.42 As a consequence, COVID-19 patients infected with the delta variant seem to have a higher risk of hospitalization, intensive care unit admission, and mortality.42 This explains why the Delta variant was the dominant strain in many countries for many months. It has also been reported that the neutralization activity against the Delta variant is strongly reduced compared to the WT, even after two doses of vaccination with mRNA vaccine.43, 44 In line with these findings, the strong cross-reaction of IgG directed WT RBD with RBD of the delta, as well as only marginal differences in avidity between IgG directed towards WT RBD and cross-reactive IgG binding to RBD delta are puzzling. However, even if the avidity of IgG binding to RBD of the delta variant is not sufficiently changed by the mutations (L432R, T478K, and P681R) inherent to RBD delta42, 45, 46 to cause a strong drop in avidity, the shift from neutral or negatively charged amino acids to positively charged amino acids due to these mutations42, 47 favors the interaction and affinity between the mutated RBD and the negatively charged ACE2 receptor. As a consequence, even at only slightly reduced avidity between RBD delta and IgG, the enhancement of binding of RBD to ACE2 can be predicted to shift the dynamics towards binding of the virus to the cells. This should result in less protective effects even of IgG of high avidity and enhances the biological effects of virus infection. Therefore, predictions on potential protective effects of high avidity IgG also needs to include the consideration of the affinity of the respective viral RBD for ACE2.
Though this pilot study with its limited number of sera will require a further extension, it already allows to conclude that the parallel determination of the quantity and avidity of IgG reacting with RBD of SARS-CoV-2 WT and omicron should allow assess the potential protective immunity towards WT, immunological related variants and the escape mutant omicron.
Thereby, the practical focus should be to determine those individuals that failed to establish high avidity IgG towards RBD of relevant variants despite vaccination, as they therefore might be of higher risk of future infection and disease.
Based on this test concept, it should be relatively easy to adjust the test system to potentially emerging additional new viral variants in the future. This would allow to asess protection towards WT virus, immunologically distinct but strongly related variants as well as immune escape variants after vaccination with mRNA derived from WT virus or any other variant. This scenario might become important as well as challenging, particularly as soon as omicron-specific vaccines will have been applied in additional vaccination programs in the future.
AUTHORS CONTRIBUTIONS
Friedhelm Struck, Eva Staschik, Karin Wochinz-Richter, Julia Maile, Erwin Soutschek, and Manfred Motz: Development, generation, and evaluation of the test system; organizing and supervising testing in-house; documentation and discussion of data; commenting and correcting the manuscript. Georg Bauer: Conceptualization; analysis of raw data; generation of the graphs, and writing the manuscript.
ACKNOWLEDGEMENTS
We thank many voluntary donors for providing sera for this study. We appreciate the valuable assistance by Jürgen Brandel (Freiburg) for the finalization of the graphs. Open Access funding enabled and organized by Projekt DEAL.
CONFLICTS OF INTEREST
E. Soutschek and M. Motz are owners of Mikrogen GmbH. E. Soutschek is the present CEO of Mikrogen GmbH. F. Struck, E. Stachik, K. Wochinz-Richter and J. Maile are employees of Mikrogen GmbH. The determination of avidity of antibodies, using immunoblots or other techniques that allow the parallel measurement of humoral immune reactions towards several antigens in one assay, has been patented by Mikrogen GmbH (WO 00/54055; PCT/EP00/01883). In addition, a new patent application for a method to determine the antibodies towards SARS-CoV-2 has been filed by Mikrogen GmbH and is pending (EP 2019/2550). Mikrogen GmbH develops and produces test systems for serological analysis of infectious diseases. G. Bauer is a member of the Medical Faculty of the University of Freiburg. He is the inventor of WO 00/54055; PCT/EP00/01883 and coinventors of EP 2019/2550.
ETHICS STATEMENT
The testing of serological parameters of SARS-CoV-2 infection in sera from hospitalized patients has been approved by the ethics commission of the Technical University of Munich (TUM) (#147/20 S-KH). Outpatients and vaccinated individuals who voluntarily took part in the serological study agreed individually and gave individual written consent to the study. The use of sera obtained from outpatients and from vaccinated individuals, for in vitro testing for antibodies towards coronaviruses was performed according to the legal specifications communicated to us by the Ethics commission of the Bavarian Medical Board [Ethik-Kommission der Bayerischen Landesärztekammer](R008-067 mat/ch), based on §24 MPG (Medizinproduktegesetz) and European norm http://eur-lex.europa.eu/legal-content/DE/TXT/?uri=OJ:L:2017:117:TOC.
STATEMENT OF PERFORMANCE ACCORDING TO RELEVANT GUIDELINES AND REGULATIONS
All research related to our study was performed in accordance with relevant guidelines and regulations. Informed consent was obtained from all participants. The use of sera given by human research participants has been performed in accordance with the Declaration of Helsinki.
Open Research
DATA AVAILABILITY STATEMENT
All raw data used for this study are presented within the study as “Grey intensity units”. Additional data are presented under Supplementary Materials