Viral load of SARS-CoV-2 Omicron is not high despite its high infectivity
Sonoka Yuasa, Jun Nakajima, and Yuna Takatsuki contributed equally to the study.
Abstract
Patients infected with the Omicron variant of severe acute respiratory syndrome coronavirus 2 has increased worldwide since the beginning of 2022 and the variant has spread more rapidly than the Delta variant, which spread in the summer of 2021. It is important to clarify the cause of the strong transmissibility of the Omicron variant to control its spread. In 694 patients with coronavirus disease 2019, the copy numbers of virus in nasopharyngeal swab-soaked samples and the viral genotypes were examined using quantitative polymerase chain reaction (PCR) and PCR-based melting curve analysis, respectively. Whole-genome sequencing was also performed to verify the viral genotyping data. There was no significant difference (p = 0.052) in the copy numbers between the Delta variant cases (median 1.5 × 105 copies/μl, n = 174) and Omicron variant cases (median 1.2 × 105 copies/μl, n = 328). During this study, Omicron BA.1 cases (median 1.1 ×105 copies/μl, n = 275) began to be replaced by BA.2 cases (median 2.3 × 105 copies/μl, n = 53), and there was no significant difference between the two groups (p = 0.33). Our results suggest that increased infectivity of the Omicron variant and its derivative BA.2 is not caused by higher viral loads but by other factors, such as increased affinity to cell receptors or immune escape.
1 INTRODUCTION
The number of patients infected with the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has increased in Tokyo as well as worldwide in January and February 2022.1, 2 It has spread more rapidly than previous variants, such as the Delta variant, which spread in Tokyo in the summer of 2021. It is important to clarify the cause of the strong transmissibility of the Omicron variant to control the spread of SARS-CoV-2.
The Tokyo Medical and Dental University (TMDU) Hospital has a role in treating severe patients with coronavirus disease 2019 (COVID-19) from all over Tokyo. This study examined the copy number and variant types of SARS-CoV-2 in nasopharyngeal swab samples from all COVID-19 patients who visited the hospital.
We previously reported that the R.1 variant (the sublineage of B.1.1.316) spread rapidly in Tokyo in March 20213 and that the copy numbers of SARS-CoV-2 in patients infected with the Delta variant were no higher than those of the R.1 and Alpha variants.4
As a possible cause of the stronger transmissibility of the Omicron variant, higher viral loads in the droplets from patients can be considered. To test this hypothesis, the precise copy numbers of viruses in the patient samples were analyzed.
2 MATERIALS AND METHODS
2.1 Patients and samples
This study used data from 694 patients who visited the TMDU hospital and were diagnosed by polymerase chain reaction (PCR) tests from July 1, 2021 to March 31, 2022. Nasopharyngeal swab samples were collected at the time of first outpatient visit or admission. The swabs were immersed in test tubes containing 1 ml of phosphate-buffered saline containing 1% dithiothreitol. This study was approved by the Medical Research Ethics Committee of TMDU (approval number: M2020-004) and was conducted in accordance with the ethical standards of the 1964 Declaration of Helsinki.
2.2 Quantitative PCR test
To detect and quantify SARS-CoV-2 in the samples, one-step reverse transcription-quantitative PCR was performed using the 2019 Novel Coronavirus Detection Kit (Shimadzu Corp.) and QuantStudio 5 Dx Real-Time PCR System (Thermo Fisher Scientific) without viral RNA purification. The copy numbers per 1 μl of sample were obtained by applying threshold cycle numbers to the calibration line of the concentration-known standard samples.
2.3 Variant screening PCR test
To determine the variant types, a melting curve analysis of the PCR products was performed. Viral RNA was purified from the swab samples using the EZ1 Virus Mini Kit v2.0 and EZ1 Advanced XL (QIAGEN). RT-PCR was performed with primers and fluorescently-labeled probes provided in the VirSNiP SARS-CoV-2 kits for Spike N501Y, E484K, E484A, L452R, and S371L/S373P (TIB Molbiol) and the LightCycler Multiplex RNA Virus Master (Roche Molecular Systems). Melting curve analysis of the PCR products was performed, and the variant types were determined by their melting temperature (Tm).
To verify the variant types determined above, whole-genome sequencing (WGS) was performed using representative samples. Libraries were prepared using the QIAseq SARS-CoV-2 Primer Panel Kit (QIAGEN) and sequenced using MiSeq (Illumina). Alignment and variant calling were performed using the CLC Genomics Workbench software (QIAGEN).
2.4 Statistical analysis
The differences in copy number and ages among the variant cases were evaluated using the Wilcoxon rank-sum test and the Steel–Dwass test for multiple comparisons.
3 RESULTS
The variant screening PCR has determined the following variant types: Alpha (501Y, 484E, 452L, and 371S/373S), Delta (501N, 484E, 452R, and 371S/373S), and Omicron (501Y, 484A, 452L, and 371L/373P). During this study, the Omicron sublineage BA.2 emerged.5 We found that analysis using the S371L/S373P probe could distinguish BA.2 (371F/373P; Tm 53.5°C) and BA.1 (371L/373P; Tm 62°C) from the other types (371S/373S; Tm 45°C).
Sequential transitions of the variants are shown in Figure 1. The Delta variant replaced the Alpha variant in August 2021 and disappeared in autumn. Subsequently, the Omicron variant emerged and spread rapidly in January and February 2022 in Tokyo. The rate of BA.2 in the Omicron cases was 1% in January, 6% in February, and 35% in March.
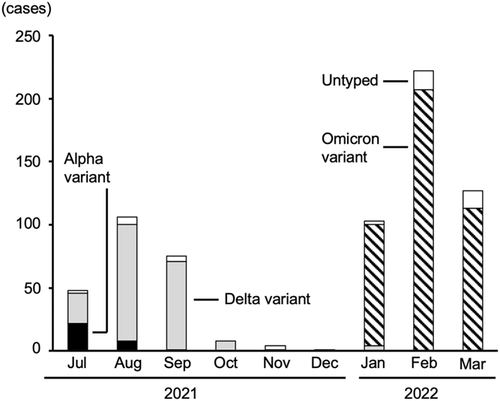
The variant types from 47 samples could not be determined because the variant screening PCR did not generate PCR products probably due to the very small copy numbers measured by quantitative PCR. Untyped samples (shown as untyped bars in Figure 1) were excluded from the analysis.
WGS was performed on approximately one-fifth of the samples and the genotyping results were consistent with those of variant screening PCR. The WGS results also showed that the majority of Omicron variants in our samples were BA1.1,6 which has the R346K mutation added to mutations in BA.1, rather than BA.1 itself, although the variant screening PCR could not distinguish BA.1 and BA1.1.
The profiles of the 502 patients infected with Delta or Omicron variants are shown in Table 1. The distribution of viral loads determined as viral copy numbers in swab-soaked samples is shown in Figure 2.
| Variant type | Delta | Omicron | ||
|---|---|---|---|---|
| Period | Total | Jan. 2022 | Feb. 2022 | Mar. 2022 |
| Case numbers | 174 | 89 | 167 | 72 |
| Age (years) mean ± SD | 49 ± 19 | 53 ± 24 | 59 ± 23 | 56 ± 25 |
| Viral loads (copies/μl) median | 1.5 × 105 | 4.9 × 105 | 6.2 × 104 | 6.4 × 104 |
- Abbreviation: SD, standard deviation.
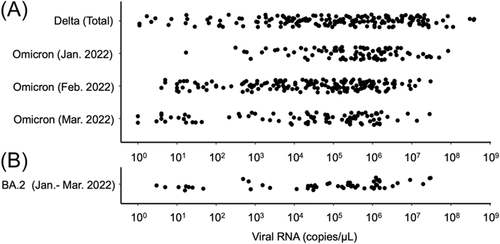
Comparing the copy numbers between the Delta cases (median 1.5 × 105 copies/μl, n = 174, 49 ± 19 years old) and Omicron variant cases (median 1.2 × 105 copies/μl, n = 328 from January to March, 57 ± 24 years old), there was no significant difference between the two groups (p = 0.052). Among the Omicron cases, the copy numbers in January were significantly higher than those in February (p = 5.4 × 10−6) and March (p = 1.6 × 10−4).
There was no significant difference (p = 0.33) between the copy numbers of the BA.1 cases (median 1.1 × 105 copies/μl, n = 275) and those of BA.2 cases (median 2.3 × 105 copies/μl, n = 53).
4 DISCUSSION
The Omicron variant is known to have higher transmissibility than previous variants.1, 2 In general, the high viral load can be one of the possible causes of increased transmissibility. As shown here, there was no significant difference in copy numbers between the Omicron cases and Delta cases in the TMDU hospital. Rather, the copy numbers of the Omicron cases appeared lower (Figure 2), although the copy numbers were widely dispersed in both cases. This finding suggests that the cause of high transmissibility of the Omicron variant is not due to high viral load, but some other undetermined factors, such as increased affinity to the receptor of human cells and decreased sensitivity to immunity due to the many mutations of the spike protein.6, 7
In March 2022, the Omicron sublineage BA.2 began to replace BA.1. There was no significant difference in copy numbers between the BA.2 cases and BA.1 cases. This suggests that a higher viral load is not the cause of rapid replacement by BA.2.
It should be noted that these results may be biased because the TMDU hospital specializes in the treatment of severe COVID-19 patients and, therefore, may not reflect all cases with COVID-19 in Tokyo. However, it is unlikely that this would affect the comparison of copy numbers among the variants, because the role of the hospital did not change throughout the study period. In fact, the sequential transitions of the numbers of each variant case at the TMDU hospital (Figure 1) were almost the same as those in Tokyo, according to a report by the Tokyo Metropolitan Institute of Public Health.8
The sequential transitions in the number of cases and the viral load of each variant can be affected by vaccination against SARS-CoV-2. According to a report by the Bureau of Social Welfare and Public Health of the Tokyo Metropolitan Government, the rate of Tokyo residents who were vaccinated three times was 4% on January 31, 21% on February 28, and 43% on March 31, 2022.9 This might be one of the reasons for the decrease in the number of cases in March in Tokyo.
This study had some limitations. The findings may have been affected by the selection bias of the patients mentioned above. The data included a small number of patients with no or mild symptoms, which was the majority of COVID-19 patients in Tokyo. However, the precise copy numbers of those cases in Tokyo were not determined. This study included some patients who were transferred to the TMDU hospital from other hospitals due to deterioration after several days of onset, which may have affected copy numbers in their samples taken at the time of admission to the hospital.
This is the first report to show a comparison of copy numbers of the Omicron variant cases and others. Our results suggest that the increased infectivity of the Omicron variant and its sublineage BA.2 is not caused by higher viral loads. Clarifying the precise mechanism of high infectivity is important for controlling the spread of the SARS-CoV-2 variant.
AUTHOR CONTRIBUTIONS
Study conception and design: Chihiro Tani-Sassa, Katsutoshi Nagano, Yumi Iwasaki, Naoya Ichimura, Yoko Nukui, and Shuji Tohda. Sample analysis: Sonoka Yuasa, Jun Nakajima, Yuna Takatsuki, Yuta Takahashi, Tomoyo Yoshimoto, and Kazunari Sonobe. WGS: Hiroaki Takeuchi, Kousuke Tanimoto, Yukie Tanaka, and Akinori Kimura. Data analysis: Naoya Ichimura and Shuji Tohda. Writing: Shuji Tohda.
ACKNOWLEDGMENTS
WGS was supported by grants JPMJCR20H2 from JST-CREST and 20nk0101612h0901 from the Japan Agency for Medical Research and Development. We would like to thank all the staff involved in COVID-19 treatment at TMDU hospital.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data and RT-PCR protocol are available from the corresponding author upon request.