Lineage analysis of human papillomavirus type 18 based on E6 region in cervical samples of Iranian women
Abstract
Distinct human papillomavirus (HPV) 18 variants are thought to differ in oncogenic potential and geographic distribution. As such, understanding the regional variants of HPV 18 would be of great importance for evolutionary, epidemiological, and biological analysis. In this regard, the sequence variations of E6 gene were investigated to characterize more common variants of HPV 18 in normal cells, premalignant, and malignant samples collected from the cervix. In total, 99 samples of HPV 18 were analyzed by polymerase chain reaction and sequencing. In overall, lineages A was identified in all study subjects, among which sublineage A4 was dominant although the difference observed was not statistically significant with regard to different stages of disease. Sublineage A4 comprised 90.9% of samples and the remaining were belonged to sublineages A1, A2, A3, and A5 at the frequency of 6.1%, 1%, 1%, and 1%, respectively. In conclusion, our findings clearly highlight the sublineage A4 of HPV 18 as the most dominant variant in Iran.
Highlights
1-First report of the HPV 18 variant analysis in Iran.
2-Lineages A are found in Iranian population.
3-All of sublineages A are found in Iranian population, but sublineage A4 are prominent.
1 INTRODUCTION
Human papillomavirus (HPV) is the most common viral sexually transmitted infection. While HPV infections generally clear up within a few months up to 2 years after acquisition, a small proportion of infections with certain HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, and 73) can persist and progress to cervical cancer, among which HPV 16 and 18 have higher chance for development of cervical cancer,1-3 as evidenced by high detection rates of HPV 16 and HPV 18 in cervical cancer at the frequency of 57% and 16%, respectively.4
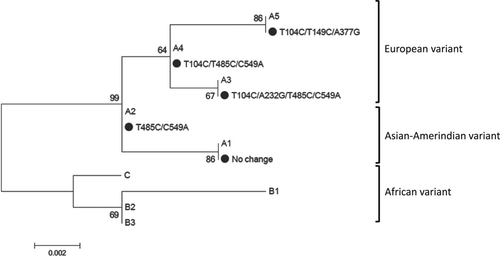
According to the definitions of HPV classification system, isolates of the same HPV type are classified as variants when nucleotide sequences of the L1 gene differ by less than 10%. When the DNA sequence of the complete viral genome is shown less than 90% to 98% similarity to other HPV genotypes is considered as a distinct lineage. A sublineage is designated when it has 0.5% to 2% nucleotide variations.5, 6 In line with these definitions, HPV 18 classified to 3 lineages and 8 distinct sublineages. HPV 18 lineages are defined as lineage A (including A1-5 sublineages), lineage B (B1-3 sublineages), and lineage C. Alternatively, sublineages of A1 and A2 were considered as Asian-Amerindian (AsAi) while A3, A4, and A5 sublineages named European (E). Moreover, B1, B2, B3, and C sublineages designated as African (Af).5
HPV 18 variants are well known to have distinctive geographical patterns, which represent the evolution of these variants linked to the population ethnicity. The interplay between HPV variants and the risk of cervical cancer progression appears to be population dependent.7
According to a meta-analysis study on the distribution of HPV types among Iranian women with invasive cervical cancer, HPV 16 is found to be the most common type in almost all different geographical regions of Iran (50.1%; 95% confidence interval [CI]: 46.4%-53.7%) followed by HPV 18 (14.4%; 95% CI: 11.9%-17%) that is consistent with most studies worldwide. Stratification of data based on histology, HPV 18 prevalence was found to be around 13.9% and 27% in squamous cell carcinoma and adenocarcinoma samples, respectively.8
While the geographical distribution of HPV types in Iran is well-documented, there is less data with regard to the HPV variants. So far, there is only one study that highlight the common HPV 16 variants in Iran.9 As such, the present study aimed to provide basic data of HPV 18 variants in Iranian women by investigating the genetic variability of HPV 18 using the complete gene of E6 (nucleotide [nt]: 105-581). The results of this study provide a rational for future studies on their evolution, pathogenicity, epidemiology, and different responses to vaccines.
2 MATERIALS AND METHODS
2.1 Study population
Ninety-nine HPV 18-positive cervical samples were included in this case-control study, which were obtained from women with normal cytology (n = 63), cervical intraepithelial lesion I-III (n = 22), and invasive cervical cancer (n = 14). Stratification by histology was revealed that thirteen of invasive cervical cancer samples (92.8%) were squamous cell carcinoma and only one sample (7.2%) was Adenocarcinoma.
ThinPrep Pap Test specimens and formalin-fixed paraffin-embedded (FFPE) tissue biopsies have been previously collected from normal and premalignant or malignant lesions, respectively.
The sample size was calculated using Fleiss formula with considering following criteria: two-sided significance level of 95%, the power of 80%, risk/prevalence difference 30%, and ratio control to case: 2. Regard to these criteria at least 32 cases and 63 controls should be evaluated.
Forty-one out of 99 samples were coinfected with other HPV types as follow: 14 samples were infected with HPV 16 and 18, 23 samples were found to be infected with HPV 18 and at least one high-risk HPV types including 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59, and four samples were coinfected with HPV 18 and a low-risk HPV types. It is worth mentioning that almost all coinfected samples were belonged to normal group or low grade precancerous lesions and only one sample of cervical cancer was coinfected with HPV 16 and 18.
Informed consent was obtained from all study subjects and the study was approved by the local ethical committee of Tehran University of Medical Sciences (98-01-27-41345). The demographic data for each case were collected from their medical records. The mean age (±SD) of women with normal, premalignant, and malignant cervix was 32.7 ± 6.8, 29.5 ± 4.5, and 50.8 ± 11.9 years, respectively.
2.2 Variant analysis of HPV 18 E6 gene
Genomic DNA of ThinPrep Pap Test specimens was extracted by High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. DNA of FFPE tissues was extracted and purified by phenol-chloroform as previously described.10
The amplification of HPV 18 E6 gene was performed using polymerase chain reaction (PCR) with the following primer pair: 5′-GATGTGAGAAACRCACCACAA-3′ and 5′-GTCGGGCTGGTAAATGTTGAT-3′ targeting an amplicon size of 662 base pair (nt 83 to 745). PCR reaction was carried out in a 50 μL reaction mixture including 100 to 200 ng of DNA template, 10 pmol of each primer, 50 μM of each dNTP, 2.5 mM of MgCl2, and 2 U of Hotstar Taq DNA polymerase (Qiagen). PCR amplification starts with an initial 5-min denaturation at 95°C, followed by 42 cycles including 95°C for 30 seconds, 55°C for 50 seconds and 72°C for 50 seconds, and a final elongation at 72°C for 5 minutes. A negative control was included in each set of PCR run.
To examine the HPV 18 variants, all the PCR products were sequenced using direct sequencing with BigDye Terminator v3.1 Cycle Sequencing Kit and a 3130 Genetic Analyzer Automated Sequencer as specified by Applied Biosystems manuals (Foster City, CA). All sequences of this study were released at http://www.ncbi.nlm.nih.gov/ with accession numbers MN563240-MN563294. To identify the lineages and sublieages of HPV 18, phylogenetic tree was constructed using the maximum likelihood method by Mega software version 6.11 The reliability was measured by calculation of bootstrap with 1000 replicates. The lineage and sublineage specific reference sequences of A1-5, B1-3, and C5 were also retrieved from GenBank database.
2.3 Statistical analysis
Statistical analysis was carried out using Fisher-exact test (Epi Info 7; Statistical Analysis System Software) and the P value less than .05 was considered as statistically significant.
3 RESULTS
The complete sequence of E6 (nt: 105-581) for 99 HPV 18-positive samples were sequenced. All sequences were compared with HPV18 E6 prototype sequence (GenBank accession number AY262282). Although all samples were belonged to lineage A, different sublineages identified, which include A1, A2, A3, A4, and A5 (Table 1 and Figure 1). Sublineages A4 were predominant at the frequency of 90.9% (90 out of 99 samples) compared with sublineages A1, A2, A3, and A5 that consist 6.1% (6 out of 99 samples), 1% (1 out of 99 samples), 1% (1 out of 99 samples), and 1% (1 out of 99 samples) of studied subjects, respectively. In overall, five distinct polymorphic patterns were identified (Table 1 and Figure 2). The maximum pairwise difference of the E6 gene sequence between any two variants was 1.9%.
| Sublineages | Acid nucleic substitutions | Amino acid changes | Nucleotide position | Studied groups | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 104 | 232 | 149 | 377 | 442 | 485 | 523 | 549 | 570 | Normal N = 63 | Premalignant N = 22 | ICC N = 14 | Total N = 99 | |||
| A1 | Prototype | T | A | T | A | A | T | C | C | A | N (%) | N (%) | N (%) | (%) | |
| A1 | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | 3 (4.7) | 2 (9.1) | (7.2) | 6 (6.1) |
| A2 | T485C/C549A | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | C | ⋯ | A | ⋯ | 0 (0.0) | 1 (4.5) | 0 (0.0) | 1 (1.0) |
| A3 | T104C*/A232G/T485C/C549A | E43G | C | G | ⋯ | ⋯ | ⋯ | C | ⋯ | A | ⋯ | 1 (1.6) | 0 (0.0) | 0 (0.0) | 1 (1.0) |
| A4 | T104C*/T485C/C549A | ⋯ | C | ⋯ | ⋯ | ⋯ | ⋯ | C | ⋯ | A | ⋯ | 58 (92.1) | 19 (86.3) | 13 (92.8) | 90 (90.9) |
| A5 | T104C*/T149C/A377G | ⋯ | C | ⋯ | C | G | ⋯ | ⋯ | ⋯ | ⋯ | ⋯ | 1 (1.6) | 0 (0.0) | 0 (0.0) | 1 (1.0) |
- Note: *Nucleotide of 104 is located at the end of long control region of HPV 18.
- Abbreviations: HPV, human papillomavirus; ICC, invasive cervical cancer.
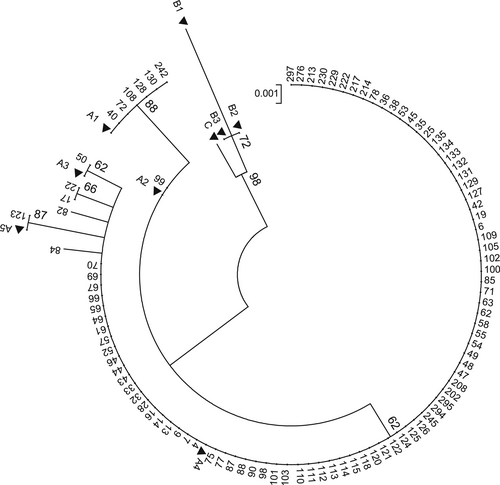
Regarding to stage of cervical samples, different sublineages were identified in study subjects. While A1 and A4 sublineages were found in cervical cancer cases, sublineages of A1, A2, and A4 were identified in premalignant group. Furthermore, sublineages of A1, A3, A4, A5, and new variants were detected in normal group (Table 1). As shown in Table 2, the difference observed between groups did not reach statistically significant (P > .05). Sublineage A4 was almost dominant in all groups, as detected in 92.1%, 86.3%, and 92.8% of normal, premalignant, and malignant groups, respectively.
| Sublineage | Studied groups | Total | P value | |
|---|---|---|---|---|
| Normal | Premalignant/malignant lesions | |||
| N (%) | N (%) | N (%) | ||
| A1 | 3 (4.7) | 3 (8.3) | 6 (6.1) | >.05 |
| A2 | 0 (0.0) | 1 (2.8) | 1 (1.0) | >.05 |
| A3 | 1 (1.6) | 0 (0.0) | 1 (1.0) | >.05 |
| A4 | 58 (92.1) | 32 (88.9) | 86 (86.9) | >.05 |
| A5 | 1 (1.6) | 0 (0.0) | 1 (1.0) | >.05 |
| Total | 63 (100) | 36 (100) | 99 (100) | |
- Abbreviation: HPV, human papillomavirus.
Sequence analysis of 99 samples showed 5 nts substitutions between positions 105 and 581 of the E6 sequence plus one substitution (T104C) at the end nt of the long control region region (Table 1). Among which, one substitution were found at the position of A232G leading to amino acid change corresponding to amino acid position E43G. The remaining four substitutions at positions of T149C, A377G, T485C, and C549A were found to be silent mutations.
4 DISCUSSION
HPV 18 variants are well known to have distinctive geographical patterns, which represent the evolution of these variants linked to the population ethnicity. The interplay between HPV variants and the risk of cervical cancer progression appears to be population dependent.7 Given the fact that HPV 18 was shown to be the second common type in Iran, no data are available with regard to the HPV 18 variants. As such, both lineages and sublineages of HPV 18 were characterized based on the sequence variations of the complete E6 gene in Iranian women with normal cervical cytology and premalignant/malignant lesions of cervix. Almost uniform distribution of HPV 18 variants was identified regardless of different groups. Distinctive sublineages were found in lineage A and, among which, sublineage A4 (European variant) was dominant. This finding is consistent with previous studies that sublineage A4 is common almost in most parts of the world, except for Sub-Saharan Africa, Eastern Asia, and Pacific where sublineages B/C are dominant.12-18 Indeed, in a study from America, 84.2% of samples belonged to lineage A (56.2% European and 43.8% AsAi variants) and the remaining of 15.8% classified to lineages B/C.15 In Netherland, lineage A was only detected as follow; European and AsAi variants were found in 71.5% and 28.5% of cases, respectively.19
Several lines of evidence have shown that distinct HPV 18 lineages exert different risks for cervical cancer progression.20-23 It is thought that African variants (lineages B and C) may have stronger oncogenic potential than AsAi variants (lineage A; sublineages A1 and A2) and European variants (lineage A; sublineages A3, A4, and A5). Epidemiological studies have shown that cervical cancer is more common in regions such as Africa where lineages B and C are frequent. However, it is worth mentioning that there is a correlation between distinct HPV 18 variants with persistency and ethnicity. While sublineage A1 was found to be more common in Eastern Asian, Pacific, South/Central Asia, North America, and South/Central America, less frequency of A2 only reported from South/Central American and South/Central Asia. Sublineages A3 and A4 are prevalent in almost all parts of the world particularly in Europe although A5 have the highest prevalence in Northern Africa. It has also been shown that sublineages B1-3 and C are common in Sub-Saharan Africa.7, 12-14, 24-27
While African variants are thought to increase the risk of cervical cancer in black women, European variants appears to have more chance for cervical cancer development in white women.7 Although yet to be documented, these findings may indicate that the immune system of white women are less capable of clearance of their European variants. The same scenario may be attributable to black women with African variants. In this regard, it has been suggested that the race-associated differences in persistency of HPV variants can lead to a less effective T-cell response in clearing the infections due to the decreased host cellular immunity.7
Different biological activities are found with regard to distinct HPV 18 variants.28 Indeed, the overexpression of cellular genes linked to the hallmarks of cancer, such as cell cycle, Wnt pathway, mTor signaling, and migration are reported to occur more effectively by E6 of African variants.23 While the lowest promoter activity was observed in European variant, AsAi variant found to exert the highest activity,29 indicating differential activities of P105 promoter among European and AsAi variants. Interestingly, epidemiological studies indicate that cervical cancer is more common in regions that the African or/and AsAi variants are more prevalent, such as Africa and Eastern Asia/Pacific.12
Regarding different sublineages among studied subjects, we found no significant difference between groups (Table 2). Although few studies have shown an association between distinctive variant and histopathology,30, 31 most studies failed to find any associations.14, 26, 32, 33 A recent study in Japan has been revealed that almost a uniform distribution of distinctive variants of HPV 18 was observed in this population as the sublineage A1 was dominant in both case and control groups.34 No significant difference also found in the HPV18 variant distribution between squamous cell carcinoma and adenocarcinoma cases.34 Although we found no significant differences when analyzing HPV 18 variants in different groups, relatively small sample size particularly positive premalignant/malignant samples should be acknowledged. Moreover, it is possible that there is an association between population-based oncogenicity of HPV 18 variants and host genetic variations, particularly HLA class I and II alleles. Indeed, it is shown that the E-G350 variant of HPV 16 had four to five fold increased risk for cancer development among Swedish women with HLA-B*44, HLA-B*51, or HLA-B*57 alleles.35 An important association between distinctive E6 variants of HPV 16 and HLA class II alleles was also found for three distinct alleles (DRB1*1501, DRB1*1502, and DQB1*0602) in Japanese women, as DRB1*1501 and DQB1*0602 alleles were significantly increased among patients with the prototype variant while DRB1*1502 was overrepresented in patients with D25E variants compared with controls.36
It has been suggested that differential coevolution of HPV 16 variants with closely related ancestral human populations might allow the dominancy of HPV16 A lineage in modern human ancestor population. This event play a key role in the different prevalence of HPV 16 variants in the world.37 In line with this fact that the geographic distribution of HPV 18 variants mirrors that found for HPV 16, it is speculated that there is an association between HPV 18 variants and ancestral human populations.34
In conclusion, our findings revealed that sublineage A4 is predominant in Iran. The predominance of A4 sublineage in this study was compatible to the sublineage distribution previously found for HPV 18 variants in Europe and America. However, further studies with larger sample size are warranted to estimate the pathogenicity risk of HPV 18 variants in Iranian population.
ACKNOWLEDGMENTS
This study has been funded and supported by Tehran University of Medical Sciences (TUMS) (grant no. 41345). It has also been part of a MSc thesis supported by Tehran University of Medical Sciences (grant no. 240/1556).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
SJ and ZS designed and directed the project. SY and ZN processed the pathological data. ZS and NH carried out experimental data and performed the analysis. SJ and SMM involved in drafting the manuscript.
ETHICS STATEMENT
Informed consent was obtained from all study subjects and the study was approved by the local ethical committee of Tehran University of Medical Sciences (98-01-27-41345).