Predominance of norovirus GI.4 from children with acute gastroenteritis in Jambi, Indonesia, 2019
The laboratory work was performed in Institute of Tropical Disease, Universitas Airlangga
Abstract
Norovirus (NoV) is one of the most important viral causes of acute gastroenteritis (AGE) in children worldwide. Only a few studies have reported AGE with NoV-positive in some cities in Indonesia. This study aimed to investigate the incidence and clinical characteristic of NoV infection, and also genotype distribution of NoV in children with AGE in Jambi, as the capital and the largest city of Jambi province, Indonesia. Stool samples were collected from children (≤15 years of age) with AGE at three participating hospitals in Jambi from February to April 2019. The detection of NoV and its genotyping were carried out by reverse-transcriptase polymerase chain reaction and direct sequencing. Of the 91 stool samples collected, 14 (15.4%) were positive for NoV. Fever, vomiting, and severe diarrhea were commonly observed in AGE with NoV, while level of dehydration was statistically significant difference between children with NoV-positive and those with NoV-negative. The most prevalent genotype was GI.4 (42.9%), followed by GII.6 (28.6%) and some other genotypes. Interestingly, this study found the predominance of GI.4, differed from previous reports in Indonesia. Continuously investigation of the circulating genotype is needed to control the NoV-infected AGE.
1 INTRODUCTION
Norovirus (NoV) is the common cause of nonbacterial acute gastroenteritis (AGE) in all age groups worldwide.1 NoV is associated with 18% of all gastroenteritis cases and causes over 200 000 deaths in developing countries annually.2 Studies on community, outpatient and hospital in developing and developed countries reported that NoV gastroenteritis accounted for 10% to 15% of severe cases in children aged less than 5 years and 9% to 15% of cases of mild to moderate diarrhea among individuals of all ages.3, 4
As the leading cause of sporadic cases and outbreaks of AGE across all age groups,5-7 NoV is highly contagious8 and mainly transmitted via fecally contaminated food or water, by direct contact with patients or vomited virus, and subsequently by contact with contaminated environmental surfaces.9 Following an incubation time of 12 to 48 hours, clinical manifestation of NoV infection starts to appear, which include nausea, vomiting, watery diarrhea, and abdominal pain.4 Diarrhea, as the most common symptom of gastroenteritis, remains a major cause of morbidity and mortality.10 In countries with effective Rotavirus vaccination programs, NoV is the most common cause of pediatric gastroenteritis requiring medical attention.11 In Indonesia, Rotavirus vaccination is included in optional immunization but not in a national immunization program,12 however, it remains unclear how much the contribution of NoV as a cause of AGE, due to the limited studies.
Norwalk viruses (NoVs) are nonenveloped, polyadenylated, single stranded, positive-sense RNA viruses of the Caliciviridae family. Based on the amino-acid sequence of VP1 (major capsid), NoVs are divided into seven genogroups (GI-GVII). The genogroup is further divided into genotypes which are defined based on either the VP1 amino-acid sequence divergence (the capsid genotype) or the nucleotide sequence divergence of the RdRp (the polymerase genotype).13, 14 NoV genogroups I, II, and IV frequently infect humans with a total of more than 30 characterized genotypes within those genogroups.15 Several studies in Indonesia reported that GII.1, GII.2, GII.4 Sydney 2012, GII.17 were detected in asymptomatic population,16 while GII.1, GII.2, GII.4 Sydney 2012, GII.6, GII.15, GII.17, GII.21, and GI.3 were detected in children less than 5 years with AGE.17 NoV circulates widely and holds considerable genetic diversity.18 Indonesia is the largest archipelago in the world, consisting of five major islands and approximately 30 smaller groups, therefore, it needs more epidemiological studies to determine the distribution of NoV genotypes that could vary geographically and longitudinally.18
In some cities in Indonesia, the incidences of NoV were reported. In Jakarta, during 1997 to 1999, NoVs were detected in 45/218 (21%) patients with acute AGE causing some symptoms including watery stool (79%), vomiting (61%), abdominal cramps (58%), and fever (50%).19 In children less than 5 years of age with AGE, 75/406 (18.47%) cases were positive for NoV and most cases clinically presented with fever (60%), diarrhea (98.7%), vomiting (88%), and some dehydration (78.7%).17 Other study in Mataram in 2017 showed that NoV was prevalent (12/38, 31.6%) in children less than 5 years of age with AGE presented with mushy stool (52.6%), vomiting (52.6%), fever (50%), severe diarrhea (84%) but no dehydration (60.5%).20 Nirwati et al17 found that genogroup GII (with genotype GII.Pe-GII.4 Sydney 2012) was predominant in patients with AGE. However, the relationship between molecular characteristics of NoV and the symptoms of its infection has not been characterized well.
Jambi is the capital and the largest city of Jambi province, located on the east coast of cental part of Sumatra island, Indonesia. About 17% of Jambi's population is concentrated in the Jambi city. Ministry of Health of Indonesia reported 54 206 diarrhea cases in children under 5 years in Jambi,21 with the prevalence about 8%,22 but the causative agents have not been reported. This study aimed to investigate the incidence and clinical characteristic of NoV infection and also genotype distribution of NoV in children with AGE in Jambi, Indonesia. The recent information about AGE with NoV in Jambi is presented for the first time, providing insight on characterization of the emerging NoV and description of clinical manifestation of its infection.
2 MATERIALS AND METHODS
2.1 Sample and data collection
Stool samples were collected from 91 children aged 3 months to 15 years (58 male, 33 female) with AGE at three participating hospitals in Jambi city, Indonesia between February and April 2019. They were collected within the first 48 hours after admission according to the World Health Organization (WHO) protocol23 and stored at −20°C before they were transported and examined in Institute of Tropical Disease, Universitas Airlangga, Surabaya, Indonesia. AGE was defined by the diarrhea (≥3 loose stools or liquid stools within a 24 hours period) and lasts no longer than 14 days, possibly accompanied by vomiting, fever, and abdominal pain. The level of dehydration were classified as follows: (a) severe dehydration, indicated by two or more of the following signs: lethargy or unconsciousness, sunken eyes, unable to drink or drinks poorly, and very slow recovery of pinched skin (≥2 seconds), (b) some dehydration, indicated by two or more of the following signs: restlessness, irritability, sunken eyes, drinks eagerly, thirsty, and slow recovery of pinched skin, and (c) no dehydration, indicated by not enough signs to classify as some or severe dehydration.24 Level of nutritional status followed WHO guidelines.25, 26 The clinical data (including data of treatment) were then used to evaluate diarrhea severity using Vesikari scoring system.27, 28 The characteristics (age, sex, nutritional status) and clinical data of the patients were retrieved from medical records. History of rotavirus vaccination was obtained from questionnaire data.
Ethical clearance for this study was obtained from the Ethics Committee of the Faculty of Medicine and Health Sciences, Universitas Jambi, Indonesia. Written informed consent was obtained by the parents or guardians of each child.
2.2 RNA extraction and reverse-transcriptase polymerase chain reaction
A 10% (wt/vol) stool suspension of each sample in distilled water was prepared by centrifuging at 15 000 rpm for 10 minutes. Then RNA was extracted from a 140 µL of the obtained supernatant using QIAamp microspin columns (QIAamp Viral RNA Mini Kit; Qiagen, Valencia, CA) to obtain a final volume of 60 µL of eluted RNA according to the manufacturer's instruction. The extracted RNA was reverse-transcribed and amplified at 65°C for 3 minutes, followed by 40°C for 1 hour using Superscript III reverse-transcriptase (Invitrogen, New York, NY) and random primers (Takara Bio, Kyoto, Japan). Polymerase chain reaction (PCR) amplification was subsequently performed to detect NoV GI and GII in the capsid gene (VP1) using the previously published primer pairs of G1SKF/R and G2SKF/R, respectively.29
PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide and visualized under UV illumination.
2.3 Sequencing and sequence analysis
Amplified complementary DNA fragments were sequenced by a direct sequencing method with the BigDye terminator cycle sequencing kit using an Applied Biosystems 3500XL Genetic Analyzer (Applied Biosystems, Foster, CA).
Genogroups and genotypes were determined using online NoV Typing Tool (http://www.rivm.nl/mpf/norovirus/typingtool). Nucleotide sequences were aligned with the reference strains by the program Molecular Evolutionary Genetic Analysis (MEGA) X (http://www.megasoftware.net). Phylogenetic trees were constructed by the Neighbor-Joining method and bootstrap resampling was performed 1000 times.
2.4 Statistical analysis
Statistical analysis was performed using IBM SPSS 22 (https://www.ibm.com/support/pages/downloading-ibm-spss-statistics-22) using the χ2 test or Fisher's exact test for categorical variables and the Mann-Whitney U test for ordinal variable. P < .05 was considered to indicate a statistically significant difference.
3 RESULTS
3.1 Virus detection rate and and characteristics of the children
Of the 91 stool samples collected, 14 (15.4%) were positive for NoV. NoVs were identified mostly in children 13 to 24 months of age group (range: 7 months to 10 years; median: 1.4 years; mean: 2.3 years) and more frequently in male patients (71.4%). The majority of the children were classified as in good nutrition status (57.1%), followed by moderate malnutrition status (21.4%), obesity (14.3%), and overweight (7.1%). All children did not have any history of rotavirus vaccination.
The age and clinical characteristics of the children with NoV-positive and those with NoV-negative are shown (Table 1). The percentage of most clinical characteristics between children with NoV-positive and those with NoV-negative was generally quite similar and not statistically significant, except the characteristic of level of dehydration. No significant difference between the two groups on age.
| Age and clinical characteristics | NoV-positive (n = 14) | NoV-negative (n = 77) | P value | OR |
|---|---|---|---|---|
| Age, y | .230 | - | ||
| < 1 | 4 (28.6%) | 25 (32.4%) | ||
| 1-2 | 8 (57.2%) | 15 (19.5%) | ||
| 2-5 | 1 (7.1%) | 21 (27.3%) | ||
| 5-15 | 1 (7.1%) | 16 (20.8%) | ||
| Fever (>38°C) | >.999 | 1.117 | ||
| Yes | 11 (78.6%) | 59 (76.6%) | ||
| No | 3 (21.4%) | 18 (23.4%) | ||
| Vomiting | .749 | 1.213 | ||
| Yes | 9 (64.3%) | 46 (59.7%) | ||
| Frequency of vomiting (episodes/d) | .198 | 0.268 | ||
| 1 | 0 (0%)a | 9 (19.6%)a | ||
| 2-4 | 7 (77.8%)a | 13 (28.2%)a | ||
| ≥5 | 2 (22.2%)a | 24 (52.2%)a | ||
| Duration of vomiting, d | .181 | 3.449 | ||
| 1 | 4 (44.4%)a | 34 (73.9%)a | ||
| 2 | 3 (33.3%)a | 4 (8.7%)a | ||
| ≥3 | 2 (22.2%)a | 8 (17.4%)a | ||
| No | 5 (35.7%) | 31 (40.3%) | ||
| Diarrhea | ||||
| Stool type | .882 | 1.300 | ||
| Watery | 5 (35.7%) | 23 (29.9%) | ||
| Mushy | 9 (64.3%) | 54 (70.1%) | ||
| Bloody | 0 (0%) | 0 (0%) | ||
| Frequency of diarrhea (times/d) | .094 | 3.056 | ||
| 3-4 | 3 (21.4%) | 35 (45.4%) | ||
| 5-9 | 7 (50.0%) | 24 (31.2%) | ||
| ≥10 | 4 (28.6%) | 18 (23.4%) | ||
| Duration of diarrhea, d | >.999 | 1.398 | ||
| 1-4 | 13 (92.9%) | 73 (94.8%) | ||
| 5 | 1 (7.1%) | 0 (0%) | ||
| ≥6 | 0 (0%) | 4 (5.2%) | ||
| Dehydration | .004b | 7.306 | ||
| No dehydration | 7 (50.0%) | 68 (88.3%) | ||
| Some dehydration | 7 (50.0%) | 8 (10.4%) | ||
| Severe dehydration | 0 (0%) | 1 (1.3%) | ||
| Vesikari score | .228 | 2.050 | ||
| Mild | 0 (0%) | 7 (9.1%) | ||
| Moderate | 5 (35.7%) | 34 (44.2%) | ||
| Severe | 9 (64.3%) | 36 (46.7%) |
- Abbreviations: AGE, acute gastroenteritis; OR, odds ratio.
- a The percentage was calculated using the number of patients with vomiting only as a denominator.
- b Statistically significant.
3.2 Genogroup and genotype of NoV and characteristics of the children
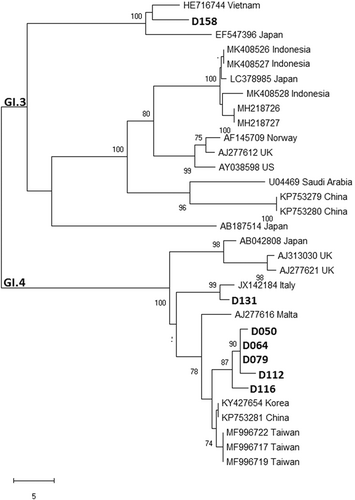
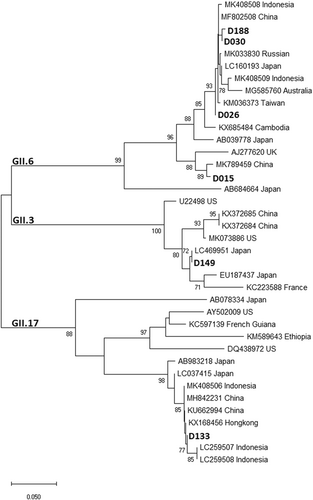
Table 2 shows all NoV genogroups and genotypes identified, along with the data of sex, age, nutritional status, and diarrhea severity of the children. Among the obtained 14 nucleotide sequences of capsid region (VP1) of NoVs, GI and GII genogrups were identified. Multiple genotypes were circulating in Jambi during the study period, with the most prevalent genotype was GI.4 (n = 6), followed by GII.6 (n = 4) and some other genotypes (GI.3, GII.3, GII.4, and GII.17) (n = 1, respectively). Five GI.4 strains obtained in this study shared 97.7 to 98.5% nucleotide (nt) sequence identity with a reference strain from Korea (KY427654), forming a cluster with strains from China and Taiwan, while the other GI.4 strain shared 98.9% nt sequence identity with a reference strain from Italy (JX142184). The four GII.6 strains shared 98.8% to 99.6% nt sequence identity with the reference strains from China (MK789459 and MF802508) and Indonesia (MK408508). The other strains with genotypes GI.3, GII.3, GII.4, GII.17 had the highest nt sequence identity (97%-100%) with the reference strains from Vietnam (HE716744), Japan (LC469951), Russia (MG892931), and Indonesia (MK408506), respectively. Most of the children had severe diarrhea, importantly, those who were infected with the predominant genotypes (GI.4 and GII.6) (67%-75%). Only three children (21.4%) were malnourished (moderate malnutrition) and all of them had severe diarrhea. Among all well-nourished (good nutritional status) children, four had severe diarrhea and they were between 1 and 2 years of age.
| Genogroup (%) | Genotype (%) | Code | Sex | Age | Nutritional status | Diarrhea severitya |
|---|---|---|---|---|---|---|
| GI (50) | GI.3 (7.1) | D158 | M | 10 mo | Good | Severe |
| GI.4 (42.9) | D050 | F | 7 mo | Good | Severe | |
| D064 | M | 1 y | Moderate malnutrition | Severe | ||
| D079 | M | 4 y | Good | Moderate | ||
| D112 | M | 10 y | Obesity | Severe | ||
| D116 | M | 18 mo | Moderate malnutrition | Severe | ||
| D131 | F | 16 mo | Good | Moderate | ||
| GII (50) | GII.3 (7.1) | D149 | M | 22 mo | Good | Moderate |
| GII.4 (7.1) | D017 | M | 17 mo | Overweight | Moderate | |
| GII.6 (28.6) | D015 | M | 13 mo | Good | Moderate | |
| D026 | M | 19 mo | Good | Severe | ||
| D030 | M | 14 m | Moderate malnutrition | Severe | ||
| D188 | F | 5 y | Obesity | Severe | ||
| GII.17 (7.1) | D133 | F | 9 mo | Good | Severe |
- Abbreviations: AGE, acute gastroenteritis; F, female; M, male.
- a Based on vesikari score.
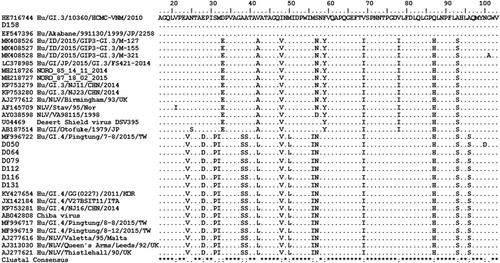
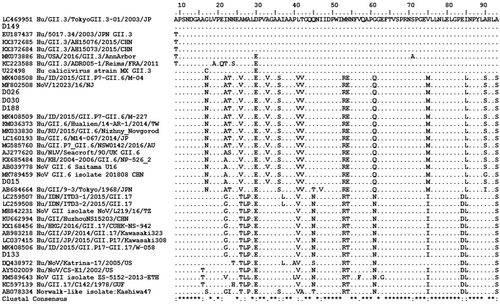
Amino-acid alignments (Figures 1 and 2) and phylogenetic trees are depicted (Figures 3 and 4), not including one strain of GII.4 due to shorter nt sequence obtained (it was repeated at least twice) than other strains.
4 DISCUSSION
In this study, the proportion of NoV in children with AGE was 15.4% in Jambi, quite similar to that in all age groups with AGE in high-mortality developing countries (14%).1 Our finding was also compatible with the reports from children and patients with all age groups in other cities (Yogyakarta, Mataram, Jakarta) in Indonesia (18.47%-31.6%).17, 19, 20 In some countries that have introduced universal rotavirus vaccination, NoV has become the leading cause of medically attended AGE in children.30 On the contrary, in Indonesia, the Rotavirus vaccination has not been implemented as a national immunization program. The high prevalence of rotavirus infection (41%-74.3%) in some cities in Indonesia (Jakarta, Bandung, Yogyakarta, Sanglah, Mataram, Bandar Lampung)17, 19, 31-33 appears to be in accordance with the lower prevalence of NoV.
Previous studies reported that NoV as a leading cause of sporadic cases and outbreaks of AGE across all age groups,5, 6 while among children, those with 13 to 24 months were prevalent.17, 34 In this study, NoV infection commonly occurred in children between 13 and 24 months of age, similar to the previous findings.
In children with AGE and NoV-positive, GI and GII have the similar proportion (50%), differed from the previous studies reported that GII NoV was the most prevalent (96%) of all sporadic AGE worldwide.13 GI.4 was the predominant circulating genotype (42.9%) in this study, however, this genotype has never been reported by previous studies in Indonesia.16, 17 GI.4 was more frequently detected in waterborne and foodborne outbreaks such as in Europian countries, Korea, and China.35-37 Those findings suggest that GI.4 can cause large outbreaks of gastroenteritis. In this study, the GI strains were isolated from children enrolled in the same hospital in the contiguous time during March 2019. It needs more investigation to determine its association with a certain outbreak and the source of its infection in Jambi.
NoV GII.6 was the second most common genotype in this study. GII.6 strains were identified in children less than 5 years of age hospitalized with AGE in other cities in Indonesia.17 In other countries, this genotype was reported to emerge such as in Japan in 2008 to 200938 and currently, it was identified among children with acute diarrhea and healthy control in Bangladesh.39 In addition, one strain each of GII.17, GI.3, and GII.3 was detected. GII.17 and GI.3 strains have been isolated in children with AGE and also in asymptomatic persons in Indonesia, but GII.3 has not.16, 17 Globally, the spread of GII.17 was identified in 13 countries across four continents40 and GI.3 was prevalent among GI strains isolated in Southern India, Australia, and New Zealand,41, 42 some were associated with outbreaks; while GII.3 was the most frequently associated with sporadic infections in infants and children in the United States.43
One strain of GII.4 was detected in this study. On the contrary, this genotype has been reported predominant for the past two decades, causing both sporadic cases and outbreaks of NoV gastroenteritis. However, minor genotypes of NoV have emerged in parts of the world, such as GII.12 in the United States in 2011, GII.13 in Nepal between 2005 and 2010, GII.14 in Japan between 2011 and 2013.44-46 The emergence and increase of the non-GII.4 genotypes may be a challenge against the candidate NoV vaccines,47 which target the globally predominant GII.4, as it is not known whether they are cross-protective against non-GII.4 strains. In this study in Jambi, only 1 GII.4 strain was identified, while GI.4 was predominant. Besides, the distribution of NoV genotypes in this study appears to be different from other regions in Indonesia.16, 17 Further investigation is warranted according to the uncommon finding. Based on data of the Manpower and Transmigration Office in Jambi province, Indonesia, hundreds of foreign workers from various countries (Japan, Korea, China, Hong Kong, and Taiwan) have been entering Jambi province in the last 3 years (2016-2019). It might contribute the spreading of the NoV genotypes to Jambi province which is shown by the close genetic relationship between the most NoV strains from Jambi and their corresponding NoV reference strains from those countries (Figures 3 and 4).
Fever (78.6%) and vomiting (64.3%) were commonly observed in AGE with NoV (Table 1). Most of childen experienced a short duration of vomiting and diarrhea. Our common findings are compatible with the previous studies,17, 19, 20, 38 but in this study, severe diarrhea was predominant. More children with NoV-positive suffered from severe diarrhea (64.3%) compared with those with NoV-negative (46.7%), it might cause a statistically significance at the level of dehydration Besides, the predominance of NoV infection was found in children 1 to 2 years, although it was not statistically significant. It confirmed the previous report that in children less than 2 years of age had a greater severity than those 2 to 4 years of age.48 The children, importantly, who were infected with the predominant NoV genotypes (GI.4 and GII.6) mostly had a severe diarrhea. It is consistent with other study reported that among genogroups GI and GII, patients infected with GI.4 and GII.6, respectively, exhibited severe symptoms with the highest Vesikari score (≥10).49 The finding of severe diarrhea in all malnourished children also agrees the previous finding of association between malnutrition and more severe NoV infections.50 Meanwhile, all well-nourished children who had severe diarrhea were between 1 and 2 years of age. The role of those factors in the severity of diarrhea due to NoV infection needs to be further investigated comprehensively.
The limitation of this study is the short monitoring period so whether or not NoV GI.4 will continue to circulate is unclear; however, it is certain that GI.4 has emerged and predominated in Jambi city and now outnumbers other genotypes of GII for the first time in Indonesia. Genotyping based on RNA-dependent RNA polymerase would enhance the completeness of NoV genotyping which is our next study plan. Since the number of NoV infections was relatively small, the statistical analysis should be confirmed further.
In conclusion, NoV was detected in 15.4% among children with AGE in Jambi, with the uncommon predominant genotype in Indonesia, GI.4. The most prevalent symptoms include fever, vomiting, and severe diarrhea. The significance of level of dehydration should be concerned. Continuously investigation of the burden and circulating genotype of NoV infection in Indonesia is needed to provide the epidemiological database for its control.
ACKNOWLEDGMENTS
This study was supported by a grant from Faculty of Medicine, Universitas Airlangga and Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from Ministry of Education, Culture, Sport, Science, and Technology in Japan, Japan Agency for Medical Research and Development (AMED: 19fm0108004). The authors are grateful to all participants who provided stool specimens and also their parents for supporting the specimen collection.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Study concept and design: Juniastuti, Wulandari. Sample collection: Wulandari. Laboratory examination: Wulandari, Wahyuni, Amin, Matondang, Dinana. Analysis and interpretation of data: Wulandari, Juniastuti, Amin, Yamani. Drafting of the manuscript: Juniastuti, Wulandari. Critical revision of the manuscript for important intellectual content: Juniastuti, Yamani, Soetjipto, Utsumi, Shoji, Lusida. Obtained funding: Juniastuti, Utsumi. Study supervision: Juniastuti.