Porphyromonas gingivalis coinfects with KSHV in oral cavities of HIV+ patients and induces viral lytic reactivation
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) infection causes several human cancers, including Kaposi's sarcoma (KS), one of the most common AIDS-associated tumors. The involvement of the oral cavity represents one common clinical manifestation of AIDS-KS individuals with periodontal diseases and an oral carriage of a variety of pathogenic bacteria, including Porphyromonas gingivalis. In the current study, we report the clinical relevance of P. gingivalis and KSHV coinfection in the oral cavity of a cohort of HIV+ patients. Furthermore, we found that P. gingivalis conditioned medium or derived lipopolysaccharide effectively induced KSHV lytic reactivation from infected oral cells. This reactivation requires TLR4 as well as the activities of p38 and Jun N-terminal kinase- mitogen-activated protein kinase signaling pathways. Our findings reveal the mechanisms through which coinfected periodontal pathogens potentially promote oncogenic virus pathogenesis in the unique niche of immunocompromised patients.
Highlights
-
Porphyromonas gingivalis co-infects with KSHV in the oral cavities of some HIV+ patients.
-
P. gingivalis induces KSHV lytic reactivation from infected oral cells through manipulation of host signaling pathways.
1 INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent of several human cancers such as Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL).1 Despite the reduced incidence of KS in the era of combined antiretroviral therapy for HIV infection, KS remains one of the most common AIDS-associated tumors and a leading cause of morbidity and mortality in this setting.2 Oral cavity involvement represents the initial manifestation of KS in 20% to 60% of HIV-associated cases.3 Oral KS lesions contain higher KSHV viral loads relative to skin KS lesions and may portend higher mortality for HIV+ patients.4 Interestingly, one recent study about oral shedding of herpesviruses in HIV+ patients has indicated that KSHV is similarly detectable across all levels of CD4 counts in these patients.5 The existing clinical data suggest that KSHV dissemination within and from the oral cavity are critical factors for KSHV infection and oral KS progression in HIV+ patients.6 It has also been noted that saliva sharing is more common in areas where KSHV is highly endemic; infection is acquired in childhood through practices such as the premastication of food for infants, candy sharing among children, and the sharing of toothbrushes.7
Periodontitis is characterized by chronic inflammation associated with oral bacteria and fungi, resulting in the destruction of periodontal ligaments and the supporting bone tissue of the tooth ultimately leading to tooth loss, high cost of oral health care, and significant morbidity.8 Several studies indicate a significantly higher prevalence of severe oral inflammation and periodontal disease for HIV+ patients.9, 10 Among those periodontal pathogens, Porphyromonas gingivalis is a well-known bacteria responsible for the development of chronic periodontitis. Published data indicate that bacteria and viruses in the oral cavity interact to facilitate periodontal disease.11 One very recent study reported impoverishment of oral microbial diversity and enrichment of specific microbiota in oral KS individuals.12 Interestingly, it has been reported that P. gingivalis can mediate epithelial cell entry of HIV-1, which represents a receptor-independent behavior.13 P. gingivalis can also induce HIV-1 in monocytes/macrophages through Toll-like receptors (TLRs).14 We recently reported that pretreatment of primary human gingival fibroblasts (HGF) and periodontal ligament fibroblasts (PDLF) with two prototypical pathogen-associated molecular patterns (PAMPs)—lipoteichoic acid (LTA) from Staphylococcus aureus and lipopolysaccharide (LPS) from P. gingivalis—increase KSHV entry and subsequent viral latent gene expression in oral cells.15 We also revealed that S. aureus derived-products, such as conditioned medium and LTA, are able to induce KSHV lytic reactivation and its coinfection with KSHV in cohort HIV+ patients.16 In this brief report, we will explore whether P. gingivalis and its products affect KSHV lytic reactivation from oral cells, how this process works, and the potential clinical relevance of these findings.
2 MATERIALS AND METHODS
2.1 Cell culture, reagents, and infection protocols
KSHV+ PEL cell line BCBL-1 was kindly provided by Dr Dean Kedes (University of Virginia) and cultured in Roswell Park Memorial Institute 1640 media with supplemented with 10% fetal bovine serum (FBS), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM l-glutamine, 0.05 mM β-mercaptoethanol, and 0.02% (wt/vol) sodium bicarbonate. Primary HGFs and PDLF were purchased from ScienCell and maintained in Dulbecco's Modified Eagle's medium supplemented with 10% FBS, 10 mM HEPES, 100 U/mL of penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B. P. gingivalis ATCC33277 and Escherichia coli ATCC25922 strains were purchased from American Type Culture Collection (ATCC), and grown as recommended by ATCC. P. gingivalis and E. coli LPS were purchased from InvivoGen, with purities more than 99.5%. SB203580 and JNK-IN-8 were purchased from Selleck Chemicals. To obtain KSHV for the infection experiments, BCBL-1 cells were incubated with 0.6 mM valproic acid for 4 to 6 days, and KSHV was purified from the culture supernatant by ultracentrifugation at 20 000g for 3 hours at 4°C. The viral pellets were resuspended in 1 of 100 of the original volume with the appropriate culture media. The infectious titers were determined as described previously.15
2.2 Patients and ethics statement
This study was approved by the Institutional Review Boards for Human Research (IRB, No. 8079) at Louisiana State University Health Science Center. All subjects were provided written informed consent. A total of 53 HIV+ patients with antiretroviral treatment in the HIV outpatient (HOP) clinic were involved. There were 21 females and 32 males in this study with an average age of 49.8 years (range, 21-67 years). The average CD4 T cell count was 607/mL (range, 31-1903/mL), and the average HIV viral loads was 6044 copies/mL (range, 24-67 082 copies/mL).
2.3 Plasma and saliva preparation
Whole blood was collected in heparin-coated tubes, and the plasma was isolated by centrifugation. The KSHV infection status was determined by using quantitative enzyme-linked immunosorbent assay (ELISA) for identifying circulating immunoglobulin G antibodies to KSHV proteins (LANA and K8.1).17, 18 To collect the whole saliva, patients rinsed with mouthwash, and saliva was collected in a wide-mouth 50 mL Nalgene tube. Typical volumes ranged between 3 and 5 mL of saliva/mouthwash. The patients were requested to not eat or smoke before providing the samples.
2.4 Enzyme-linked immunosorbent assay
The concentrations of total bacterial LPS in saliva were quantified using LPS ELISA Kit (Cloud-Clone) as recommended by the manufacturer.
2.5 RNA interference and quantitative real-time polymerase chain reaction/polymerase chain reaction
For RNA interference (RNAi) assays, TLR4 ON-TARGET plus SMART pool siRNA (Dharmacon), or negative control siRNA were delivered using the DharmaFECT transfection reagent. Total RNA was isolated using the RNeasy Mini Kit (Qiagen), and complementary DNA was synthesized using a SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen). Primers used for amplification of target genes were listed in Table S1. Amplification was carried out using an iCycler iQ Real-Time PCR Detection System, and cycle threshold (Ct) values were tabulated in duplicate for each gene of interest in each experiment. “No template” (water) controls were used to ensure minimal background contamination. Using mean Ct values tabulated for each gene, and paired Ct values for β-actin as a loading control, fold changes for experimental groups relative to assigned controls were calculated using automated iQ5 2.0 software (Bio-rad). When polymerase chain reaction (PCR) was used to detect bacteria and viruses in saliva samples, the positive (DNAs from P. gingivalis ATCC33277 or KSHV+ BCBL-1 cells) and negative (water) template controls were used too.
2.6 Immunoblotting
Total cell lysates (30 µg) were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated with 100 to 200 µg/mL of antibodies for TLR4, phosphor- or total-p38, phosphor- or total-JNK, β-Actin (Cell Signaling), and KSHV-RTA (Abbiotec).
2.7 Statistical analysis
Significance for differences between experimental and control groups was determined using the two-tailed Student t test (Excel 2016).
3 RESULTS AND DISCUSSION
3.1 Clinical prevalence of P. gingivalis and KSHV shedding within the oral cavity of cohort HIV+ patients
In saliva samples obtained from 53 cohort HIV+ patients, we found that 41.5% (22/53) were P. gingivalis positive, and 18.9% (10/53) were KSHV-Lana positive (a viral protein marker representing latent infection) (Figure 1A,B). Notably, six patients (11.3%) were P. gingivalis/KSHV double-positive, providing clinical evidence for the coinfection of this specific periodontal bacterial species and oncogenic virus in the oral cavity of HIV+ patients. We noticed that 41.5% of the HIV+ patients (22/53) were KSHV seropositive, which is much higher than determined from their saliva samples (18.9%); this discrepancy is likely due to the difficultly of acquiring KSHV-infected oral cells in just one saliva collection. Therefore, we speculate that the actual rates of oral KSHV positive as well as P. gingivalis/KSHV double-positive cases are potentially higher than what we found. Currently, there are no commercial kits available to measure the LPS concentration derived from a single bacteria species such as P. gingivalis, thus we can only measure total bacterial LPS levels. Our data indicate that HIV+/KSHV+ patients have higher levels of total salivary LPS than HIV+/KSHV− patients, indicating the potential clinical implication of periodontal bacterial PAMPs in oral KSHV pathogenesis and related disease progression (Figure 1C).
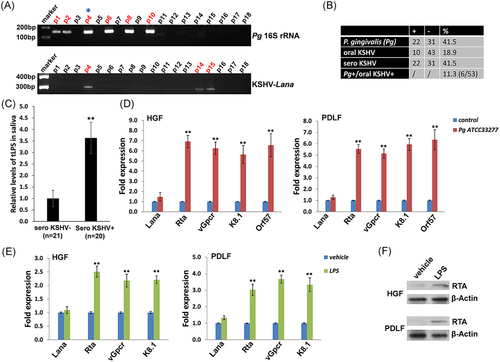
3.2 Induction of KSHV lytic reactivation from infected oral cells by P. gingivalis culture and components
We found that treating latently infected primary human oral fibroblasts (HGF and PDLF) with filtered conditioned medium from P. gingivalis ATCC33277 strain-induced significantly higher expression of KSHV lytic genes, including Rta, vGpcr, K8.1, and Orf57, when compared to cells treated with a fresh medium control (Figure 1D). Moreover, these inducible effects were also observed in P. gingivalis-derived LPS-treated oral cells with virus infection (Figure 1E), although to a lesser extent. Furthermore, immunoblot results confirmed the induction of RTA expression from P. gingivalis-derived LPS-treated oral cells (Figure 1F). However, we did not observe similar effects using conditioned medium or purified LPS from other nonperiodontal Gram-negative bacteria such as E. coli (Figure S1), indicating specificity for bacterial species. In fact, P. gingivalis-derived LPS exhibits unique features compared to the LPS of other species, including differences in the structure of the O-antigen, as well as in the acylation patterns and receptor-activating capacities of the lipid A component.19, 20 Interestingly, one recent study reported that P. gingivalis LPS and E. coli LPS differently regulated cytokine production in HGFs.21 Another study showed that whole blood cell cultures populations obtained from healthy and chronic periodontitis patients may differ in the cytokine response to P. gingivalis LPS but not E. coli LPS.22
3.3 TLR4 and intracellular signaling pathways are required for the induction of KSHV lytic reactivation by P. gingivalis conditioned medium and LPS
The receptor for P. gingivalis LPS has initially been reported as TLR2, although this observation remains controversial because synthetic P. gingivalis lipid A activates TLR4 but not TLR2,23 thus the TLR2 activity of P. gingivalis LPS might be attributed to a contaminant lipoprotein.24 Moreover, a recent study states that P. gingivalis LPS activity is mediated exclusively through TLR4.25 Interestingly, one recent study demonstrated the role of TLR4-mediated inflammation in KSHV-induced tumorigenesis.26 In this study, we first assessed TLR4 expression in oral cells, using human monocyte THP-1 as a positive control. Our real-time PCR data indicated that TLR4 is expressed in oral fibroblasts, HGF, and PDLF (Figure 2A). Silencing of TLR4 by RNAi significantly impaired KSHV lytic gene expression (eg, Rta, a viral lytic reactivation activator), induced by P. gingivalis conditioned medium and/or LPS (Figure 2B-D). However, in contrast to near-complete impairment of viral lytic gene expression induced by LPS, silencing of TLR4 had only partial blocking effects on viral lytic gene expression induced by P. gingivalis conditioned medium (Figure 2C,D). These results indicate that additional bacterial products besides LPS may induce viral lytic gene expression, such as the short-chain fatty acids reported previously.27

P. gingivalis infection has been found to activate several intracellular signaling pathways such as p38 and Jun N-terminal protein kinase (JNK)-mitogen-activated protein kinase pathways.28 Interestingly, the activities of p38 and JNK pathways have been found to be closely related to KSHV lytic reactivation.29 Here, we demonstrated that both P. gingivalis conditioned medium and LPS effectively increased the phosphorylation of p38 and JNK proteins from KSHV-infected oral cells (Figure 2E). Pretreatment of specific p38 inhibitor (SB203580) or JNK inhibitor (JNK-IN-8) significantly reduced viral lytic gene expression induced by P. gingivalis conditioned medium and/or LPS (Figure 2F,G). Again, we noticed that blocking these two signaling pathways did not completely hinder viral lytic reactivation induced by P. gingivalis conditioned medium (Figure 2F), implying that there are additional mechanisms involved in this induction.
In summary, our findings demonstrate the clinical relevance of P. gingivalis and KSHV coinfection in the oral cavity of cohort HIV+ patients, which (including the whole bacteria or bacterial products) may facilitate oncogenic virus lytic reactivation and dissemination.
ACKNOWLEDGMENTS
This study was supported by NIH/NCI R01CA228166. This study was also supported in part by the Arkansas Bioscience Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.