Development of a sensitive and specific nanoparticle-assisted PCR assay for detecting HPV-16 and HPV-18 DNA
Abstract
Carcinoma precursor lesion caused by persistent infection of human papillomavirus (HPV) types 16 and 18 is known as a principal inducer of cervical cancer. Therefore, rapid and effective detection of HPV-16 and HPV-18 infection at early stage is an important strategy for preventing such disease. In this study, a novel duplex nanoparticle-assisted polymerase chain reaction (nanoPCR) assay was developed to detect both of the two genotypes simultaneously. Two pairs of primers for nanoPCR were designed based on the conserved region within the early 6 (E6) gene of HPV-16 and HPV-18, respectively. After optimizing reaction conditions, the nanoPCR assay displayed 10-fold more sensitive than that of conventional PCR and showed high specificity. The detection limit of nanoPCR was 1.7 × 101 copies/μL for HPV-16, 1.2 × 102 copies/μL for HPV-18, and no cross-reaction was detected after using other viruses or HPV subtypes as templates. Of 209 clinical samples collected from patients, as also confirmed by sequencing, the nanoPCR method gave consistent results with conventional PCR assay: 7 positives for HPV-16, 4 positives for HPV-18, and no co-infection. Here is the first report to introduce a reproducible nanoPCR assay for detecting HPV DNA with high sensitivity and specificity, which may point out a useful diagnostic tool for potential clinical application.
Highlights
-
A novel nanoPCR method was developed for the detection of HPV-16 and HPV-18.
-
The HPV nanoPCR assay was 10-fold more sensitive than conventional PCR assay.
-
The lower detection limit was 1.7 × 101 copies/μL for HPV-16 and 1.2 × 102 copies/μL for HPV-18.
1 INTRODUCTION
Human papillomavirus (HPV), belonging to the papillomavirus family, is a small double-stranded circular DNA virus with a proximate length of 8000 bp.1 The molecular structure of HPV contains a long control (LC) region, early (E) regions, and late (L) regions. The E regions, including E1, E2, E4, E5, E6, and E7, are involved in controlling viral replication, transcription, and translation, whereas L regions contain a L2 and a highly conserved L1, participating in the regulation of capsid proteins encoding and viral invasion.2 Normally, sexually active and indirect contact are major propagation ways of HPV transmission, which normally occurs in epidermis and mucosal tissues.3 To date, more than 170 genotypes of HPV have been identified and are categorized as high-, medium-, and low-risk according to their biological characters and oncogenic potential. Notably, constant high-risk HPV (HR-HPV) infection, in particular, HPV-16 and HPV-18 subtypes, has been demonstrated as key players in inducing cervical carcinoma, which is the second common cancer in women among the world.4
Theoretically, after infection, the HPV maintains its proliferation and protein synthesis depending on the replication of host cells after integrating its DNA into the host genome.5 Indeed, the open-reading frames of E1 and E2 are broken during DNA recombination, thus releasing the E2-repressed E6 and E7 oncogenic proteins expression.6 The function of E6 has been verified to promote cell proliferation by accelerating degradation of the tumor suppressor p53 protein as well as suppressing its downstream p21 (a cell cycle inhibitor) expression.7 Whereas E7 represses the retinoblastoma phosphorylation, resulting in the activation of transcriptional regulator E2F, thus establishing continuous cell cycle progression and ultimately leading to malignant proliferating of host cells.2, 8 Hence, the E6 and E7 oncoproteins play crucial roles in persistent infection of HPV.
Although it takes several decades to develop cervical carcinoma lesions after carrying HPV, the morbidity and mortality of cervical cancer (CC) are continuously increasing, especially in developing countries such as China, the reason of which may mainly due to the low efficiency of early detection and diagnosis of HPV.9 Therefore, it is of great significance to establish effective diagnostic tools for early HPV detection. Currently, almost all of HPV detection methods are proceeded based on the determination of virus DNA, among which the most common one is amplification of target fragments with specific primers using polymerase chain reaction (PCR).10 The PCR technique, based on the amplification of conserved regions of HPV, has developed from primary IU/IWDO, MY09/11, and GP5/6 to the highly sensitive SPF10 system and later PCR reverse dot blot, until quantitative PCR (qPCR) derived Cobas4800 strategies.11, 12 However, application of these methods is either limited by the sensitivity to detect latent infection, time-consuming, or requirement of expensive equipment and professional operations. In addition to PCR, other detection technologies have also been introduced, including hybrid capture 2,13 loop-mediated isothermal amplification (LAMP),14 and E6/E7 messenger RNA detection.15 Advantages exist whereas shortcomings remain to be addressed, such as complicated experimental conditions and easily contaminated samples.
In this study, a novel sensitive and specific duplex nanoparticle-assisted PCR (nanoPCR) assay was first described to quickly detect E6 gene of both HPV-16 and HPV-18 genotypes, respectively. NanoPCR system is advanced for its containing of solid gold nanometal particles (1-100 nm).16 These nanofluids allow the reaction reach target temperature more quickly, thus shortening the nontargeted time period, reducing nonspecific amplifications and thereby improving the reaction efficiency.17 The nanoPCR assay has been applied to many animal virus detections due to its high sensitivity and specificity, as well as easier and faster performance.18 Hence the assay developed here is of strong valuable for potential clinical application of HPV early diagnostic detection.
2 MATERIALS AND METHODS
2.1 Viruses and recombinant plasmids
The following viruses and HPV subtypes used to evaluate the specificity of HPV nanoPCR were obtained from the Central Laboratory of The Affiliated Hospital of Yangzhou University, Yangzhou, China: hepatitis B virus (HBV), hepatitis C virus (HCV), human immunodeficiency virus (HIV), HPV-11, HPV-16, HPV-18, HPV-31, HPV-39, and HPV-52. The recombinant plasmids pUC57-HPV-16-E6 and pUC57-HPV-18-E6, containing complete coding sequences of E6 gene of HPV-16 or HPV-18, were purchased from Sangon Biotech (Shanghai, China) and amplified in Escherichia coli DH5α then purified in accordance with the TIANprep Mini Plasmid Kit (Beijing Tiangen Biotech Company, Beijing, China). The plasmids were stored at −20°C and used as standards.
2.2 Primers design
The full-length sequences of E6 gene of both HPV-16 and HPV-18 were obtained from GenBank (GenBank accession numbers: NC_001526.4 and NC_001357.1). Two pairs of primers specifically for HPV-16-E6 and HPV-18-E6, with amplicon size of 383 and 259 bp, respectively, were designed using PrimerQuest (https://sg.idtdna.com/pages/tools/primerquest). Primers were synthesized by Sangon Biotech and their sequences are listed (Table 1).
| HPV subtype | Gene | Primer | Primer sequence (5′-3′) | Length (bp) |
|---|---|---|---|---|
| HPV-16 | E6 | F | CACAGAGCTGCAAACAACTATAC | 383 |
| R | TTGATGATCTGCAACAAGACATAC | |||
| HPV-18 | E6 | F | ACACTTCACTGCAAGACATAGA | 259 |
| R | CACCGCAGGCACCTTATTA |
- Abbreviations: HPV, human papillomavirus; nanoPCR, nanoparticle-assisted polymerase chain reaction.
2.3 Clinical specimens and DNA preparation
A total of 209 random chosen de-identified cervical scrape specimens for early diagnosis of HPV infection collected from gynecological clinic patients in 2019 were kept in the Central Laboratory of The Affiliated Hospital of Yangzhou University. This study was approved by the Ethics Committee of The Affiliated Hospital of Yangzhou University. DNA of the clinical samples and DNA or RNA of above reference viruses were extracted with a TIANamp virus genomic DNA/RNA kit (Beijing Tiangen Biotech Company) according to the manufacturer's instructions. RNA was then reverse-transcribed to complementary DNA (cDNA) using the TranScript Firststrand cDNA Synthesis SuperMix (Beijing TransGen Biotech Company). The extracted DNA or synthesized cDNA were eluted in RNase-free ddH2O and stored at −20℃.
2.4 Optimization of the HPV nanoPCR assay
The nanoPCR Kit (NPK02) was purchased from GREDBIO (Weihai, China). Independent experiments were performed to optimize the annealing temperature, primers, and standard plasmids concentrations for the HPV nanoPCR assay. Specifically, the reaction was performed in a 12 μL system in the PCR Express Thermo Cycler (Thermo Hybaid); the annealing temperatures were tested from 50℃ to 60℃ by 1℃ gradient increasing; each pair of forward and reverse primers (20 μM) was tested in volumes ranging from 0.1 to 1 μL with 0.1 μL increasing and templates volume (mix of 1.7 × 109 copies/μL for HPV-16-E6 and 1.2 × 1010 copies/μL for HPV-18-E6) was raised from 0.2 to 1.6 μL in increments of 0.2 μL. PCR products were visualized by electrophoresis on 1.5% agarose gels and read by iBright CL1500 Imaging System (Thermo Fisher Scientific).
2.5 Conventional HPV PCR assay
The conventional PCR kit (2X Taq PCR Mastermix) was purchased from Beijing Tiangen Biotech Company. The conventional HPV PCR assay was performed in a 12 μL system based on the optimized HPV nanoPCR condition including 1.0 μL of mixed HPV-16-E6 and HPV-18-E6 standard plasmids or DNA from clinical samples, 0.6 μL of each of forward and reverse primers (20 μM) for HPV-16-E6 and HPV-18-E6, 6.0 μL of 2X PCR buffer, and ultimately adding ddH2O up to 12 μL. The conventional PCR assay was conducted following the same protocol as the HPV nanoPCR assay.
2.6 Sensitivity and specificity of HPV nanoPCR assay
To determine the sensitivity of the HPV nanoPCR assay, the HPV-16-E6 and HPV-18-E6 plasmids were quantified using NanoDrop One Spectrophotometer (Thermo Fisher Scientific) (1.7 × 109 copies/μL for HPV-16-E6 and 1.2 × 1010 copies/μL for HPV-18-E6, respectively). The sensitivity of HPV nanoPCR assay was accessed through comparison with that of conventional PCR assay by using serially 10-fold diluted plasmids: 1.7 × 109 to 1.7 × 100 copies/μL for HPV-16-E6 and 1.2 × 1010 to 1.2 × 101 copies/μL for HPV-18-E6, respectively. Of note, each diluted sample was tested as a template using the optimized reaction parameters and ddH2O was applied as negative control. Amplified products were subjected to electrophoresis on 1.5% agarose gels.
To evaluate the specificity of the HPV nanoPCR assay, DNA, or cDNA of the following viruses and HPV subtypes were separately subjected to the established HPV nanoPCR assay: HBV, HCV, HIV, HPV-11, HPV-16, HPV-18, HPV-31, HPV-39, and HPV-52. The HPV-16-E6 and HPV-18-E6 plasmids were used as positive controls and ddH2O as negative control. PCR products were analyzed by electrophoresis on 1.5% agarose gels.
2.7 Detection of HPV in clinical samples
DNA of all 209 samples were prepared as described in Section 2.3 and were subjected to the HPV nanoPCR and conventional PCR assays. Amplicons from samples which were recognized as either HPV-16 or HPV-18 positive by the nanoPCR method were subsequently sent to Sangon Biotech for sequencing. The GenBank BLAST searching tool was introduced to determine the consistency between the sequenced amplicons and the accessed HPV-E6 sequences.
3 RESULTS
3.1 Optimization of the HPV nanoPCR conditions
The recombinant plasmids pUC57-HPV-16-E6 and pUC57-HPV-18-E6 were applied as templates to optimize HPV nanoPCR conditions. The annealing temperatures were primarily tested from 50℃ to 60℃ by gradient 1℃ increasing, and the best amplification point was found at 56℃. Afterwards, volume of primers was chosen at 0.6 μL for each of forward and reverse primers (20 μM) based on the testing from 0.1 to 1 μL using 56℃ annealing temperature. Lastly, volume of the mixed plasmid DNA in the nanoPCR system was chosen at 1.0 μL after testing from 0.2 to 1.6 μL in increments of 0.2 μL under 56℃ annealing temperature and 0.6 μL of each of forward and reverse primers.
After optimizing, the HPV nanoPCR assay in a 12 μL reaction mixture was finally established as 1.0 μL of extracted DNA or mix of standard plasmids, 0.6 μL of each of forward and reverse primers (20 μM) for HPV-16-E6 and HPV-18-E6, 6.0 μL of 2X nanoPCR buffer, and 0.5 μL of Taq DNA polymerase (5 U/μL); and ultimately adding ddH2O up to 12 μL. The nanoPCR reaction condition was 5 minutes at 94℃; followed by 30 cycles of 94℃ for 30 seconds, 56℃ for 30 seconds, and 72℃ for 50 seconds; and a final elongation at 72℃ for 10 minutes.
3.2 Sensitivity of the HPV nanoPCR assay
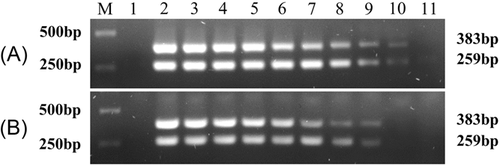
The sensitivity of the HPV nanoPCR and conventional PCR assays was compared using a series of diluted pUC57-HPV-16-E6 and pUC57-HPV-18-E6 templates as indicated in Section 2.6. The detection limit for pUC57-HPV-16-E6 was determined as 1.7 × 101 copies/μL with the nanoPCR assay (Figure 1A), whereas it was 1.7 × 102 copies/μL with the conventional PCR assay (Figure 1B). Besides, for pUC57-HPV-18-E6 template, the lowest detection concentration was 1.2 × 102 copies/μL with the nanoPCR assay (Figure 1A), while it was 1.2 × 103 copies/μL with the conventional PCR assay (Figure 1B). These data indicated that the HPV nanoPCR assay was 10 times more sensitive than that of the conventional PCR assay.
3.3 Specificity of the HPV nanoPCR assay
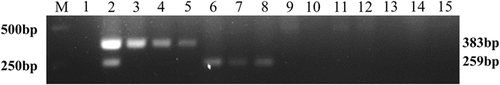
The specificity of HPV-16 and HPV-18 nanoPCR assay was accessed, as described in Section 2.6, by testing the DNA or cDNA of three different viruses and six HPV subtypes using HPV-16-E6 and HPV-18-E6 primers. Results showed the optimized HPV nanoPCR assay amplified all of HPV-16 and HPV-18 subtypes but none amplicons appeared after adding other DNA or cDNA as templates (Figure 2). These data indicated that the established HPV-16 and HPV-18 nanoPCR assay was specific.
3.4 Detection of HPV-16 and HPV-18 in clinical specimens
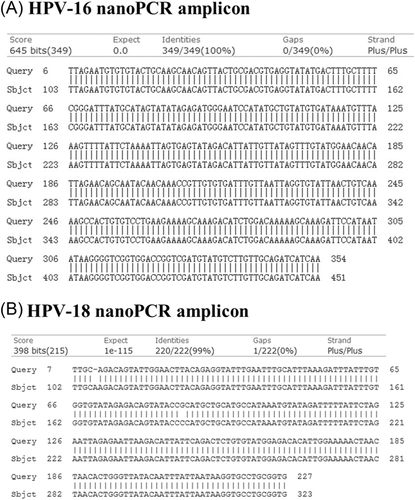
A total of 209 clinical specimens were processed by the HPV nanoPCR and conventional PCR assays. According to the results from both methods, the nanoPCR assay gave consistent outcomes with conventional PCR assay: seven positives for HPV-16, four positives for HPV-18, and no co-infection detected. The detection rates were 3.35% for HPV-16 and 1.91% for HPV-18, respectively. The amplicons obtained from HPV nanoPCR assay were subsequently sent to sequence. Results confirmed that the nanoPCR assay correctly identified HPV-16 and HPV-18 (Figure 3) after aligning with GeneBank reference sequences, as indicated in Section 2.2.
4 DISCUSSION
It has been reported that the majority of sexually active women would be infected by at least one HPV subtype during lifetime, but normally most of them maintain in subclinical status and the infection could be subsequently cleaned by immune system.19 However, some of the infections, in particular those HR-HPV subtypes, persist and evolve from low to high grade of cervical intraepithelial neoplasia over decades, and eventually the infiltration converts to CC. In fact, HPV-16 and HPV-18 have been demonstrated as the most prevalent HR-HPV genotypes in women worldwide.2, 20 Therefore, early, effective and regular diagnosis of HPV infection is emerging to be an important tool for preventing CC. In addition, males cannot sit back to relax as several subtypes, such as low-risk HPV-6 and HPV-11, cause condyloma acuminatum in men.21
At present, PCR method to identify HPV DNA has been regarded as a golden standard for detecting HPV infection,11, 22 and thus the majority of commercial HPV diagnostic kits are developed based on PCR strategies. Moreover, the HPV suspension array has been achieved currently through combing PCR technique (to amplify conserved regions of different subtypes) with reverse cross-blot hybridization (to reverse hybridize amplicons with coated specific probes). In addition, qPCR, LAMP, as well as hybrid capture 2 assays have also been established and regarded as useful tools for HPV detection.23 However, due to either the limitation of detection by conventional PCR, requirement of expensive equipment by qPCR, or easily contaminated by LAMP, other easier and more sensitive methods are still needed.
In this study, a nanoPCR assay to detect HPV was firstly optimized and developed. Because of containing high thermal conductive nanofluids, the nanoPCR assay has been proved to be extremely specific and more sensitive, which are essential characters in case of monitoring low virus concentrations, than conventional PCR assay in detecting diverse viruses.24 For example, nanoPCR method has been introduced to be of high specific and 100-1000 times more sensitive than routine PCR in diagnosing certain viruses including pseudorabies virus, porcine bocavirus, mink enteritis virus, bovine respiratory syncytial virus as well as brain-eating amoebae.25-27
The nanoPCR assay established here that targets the E6 gene of both HPV-16 and HPV-18 subtypes is an effective and reagent-saving reaction (12 μL system). Additionally, although detection of clinical specimens gave same results with nanoPCR and current commercial PCR strategies, the former assay was confirmed 10-times more sensitive than the later one via amplifying diluted standards plasmids. Besides, the HPV nanoPCR assay was specific as it did not amplify three other viruses or four else HPV subtypes. Furthermore, as other HR-HPV genotypes than HPV-16 and HPV-18 are also crucial in causing CC, such nanoPCR detecting strategies can be extended for commercial diagnosis of other cancer related HR- or low-risk-HPV genotypes. Therefore, the method introduced here may be benefit for early HPV detection, thus contributing to minimize the morbidity of CC.
In conclusion, an easy, reagent-saving, sensitive, and specific nanoPCR assay was described here for detecting HPV-16 and HPV-18 subtypes simultaneously. This novel assay should be useful for both clinical HPV type diagnosing and for potential studying of HPV epidemiology and pathology.
ACKNOWLEDGMENTS
This study was supported by the Research Foundation for Advanced Scholars in the Affiliated Hospital of Yangzhou University, Yangzhou, China (2019MXJ).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.