Quasispecies dynamics of a hepatitis E virus 4 from the feces and liver biopsy of an acute hepatitis E patient during virus clearance
Abstract
The presence of a quasispecies in hepatitis E virus (HEV) infection has been documented, however, the implications of a quasispecies in HEV-host interaction are poorly understood. Here, we analyzed the whole genome sequences of a HEV 4d from the feces and liver biopsy of a patient during the icteric and convalescent phases in an acute hepatitis E infection. Viral RNAs were extracted, reversely transcribed and seven fragments encompassing the entire viral genome were amplified and cloned. By sequencing multiple colonies of each cloned viral genome amplicon with Sanger sequencing, we verified the existence of the HEV quasispecies or intra-host genetic variations within the fecal and liver biopsy samples. There were broader genetic variations in the HEV ORF1 region including the PCP, HPX, and RdRp regions during the convalescent phase whereas more genetic variations in the ORF2 P domain during the icteric phase. The quasispecies dynamics might reflect host immune pressure during viral clearance.
Highlights
-
Verify the presence of viral quasispecies in an acute hepatitis E virus (HEV) infection.
-
Different pattern of quasispecies during the icteric phase and convalescent phase.
-
Identification of hot spot genetic variations in the P domain of ORF2.
1 INTRODUCTION
Hepatitis E virus (HEV) is a Hepeviridae family member. HEV usually causes acute infection and causes chronic viral hepatitis in immunocompromised individuals. The HEV genome is a 7.2-kb positive-sense RNA that encodes three open reading frames (ORFs). ORF1 encodes viral nonstructural proteins that participate in viral replication. ORF1 is composed of the methyltransferase domain (Met), Y domain (Y), papain-like cysteine protease (PCP), hypervariable region, proline-rich domain, X domain (HPX), helicase (Hel) and RNA-dependent RNA polymerase (RdRp). ORF2 encodes capsid protein that assembles the virion. ORF2 encodes a viral capsid protein of 660 amino acids. The capsid protein consists of the S domain (aa 118-313), M domain (aa 314-453), and the P domain (aa 454-606) that forms the protrusion projecting from the viral shell.1 Recently, it was reported that an alternative translation initiation at ORF2 results in the secreted ORF2 (ORF2s). The glycosylated ORF2s without the putative cell-receptor-binding peptide inhibits antibody-mediated neutralization.2 ORF3 encodes a small multifunctional protein that is involved in virion release.3 HEV is mainly transmitted through a fecal-oral route. There are at least four HEV genotypes (genotypes 1, 2, 3, and 4) that can infect humans in the genus of Hepevirus. Genotype 1 and 2 mainly infect humans. Genotypes 3 and 4 are zoonotic and animals such as pigs serve as reservoirs.4
The RNA-dependent RNA polymerase of most RNA viruses lack proofreading and undergo error-prone replication that results in viral genomic diversity. During chronic viral infection such as hepatitis C, long-term virus error-prone replication results in a viral population with a high degree of sequence variations within an infected individual, namely viral quasispecies.5 The presence of quasispecies or intra-host populations and some genetic variants in low-frequency impact host immune responses during viral infection6 and may have important clinical implications.7 There are pieces of evidence for HEV quasispecies in acute and chronic HEV infection that imply the influence of the viral genetic heterogeneity in viral persistence.8, 9
In this study, we analyzed the entire genome sequence of a HEV4 from the fecal sample during the icteric phase and the liver biopsy sample during the convalescent phase of a patient with an acute hepatitis E infection. By sequencing multiple colonies of cloned amplicons with Sanger sequencing, we verified the existence of HEV quasispecies within the fecal and liver biopsy samples and found different quasispecies patterns during the icteric phase and the convalescent phase.
2 MATERIALS AND METHODS
2.1 Ethic and patient
This study was reviewed and approved by the Ethics Committee of Shanghai Public Health Clinical Center and the School of Basic Medical Sciences, Shanghai Medical College, Fudan University. The procedures were carried out in accordance with approved guidelines. Informed consent was obtained from the subject. The liver function and the serologic status of the patient were routinely tested by the department of diagnosis in the hospital. The serologic status was analyzed by enzyme-linked immunosorbent assay (ELISA). The cutoff for IgG (S/CO) is 1.0 and the cutoff for IgM (S/CO) is 1.2.
2.2 RNA extraction and reverse transcription
Fecal samples were first diluted in phosphate buffered saline, and clarified by full-speed centrifugation for 10 minutes twice. The RNAs in the supernatants were extracted by using a QIAamp Viral RNA Mini kit (QIAgen) and eluted in RNase-free H2O. Similarly, RNAs from plasma were extracted by using a QIAamp Viral RNA Mini kit and eluted in RNase-free H2O. RNAs from the liver biopsy were extracted with TRIzol LS (Invitrogen) and dissolved in RNase-free H2O. Reverse transcription was performed with Supercript II (Invitrogen). Briefly, 8 μL of RNA samples was reversely transcribed with 1 μL random primer (dN6), according to the manufacturer's protocol.
2.3 Quantification of HEV RNA by real-time PCR
HEV RNA was quantified by real-time PCR (R-PCR) with a TaqMan-based method. Briefly, 2.5 μL of RNAs was reversely transcribed and PCR amplified by using a Takara One Step PrimeScript RT-PCR Kit in a 25 μL reaction mixture as follows: 2 × one-step RT-PCR buffer, 12.5 μL; EX Taq HS, 0.5 μL; RT Enzyme, 0.5 μL; primer mix (10μM each), 1.6 μL; probe (10 μM), 0.4 μL; H2O up to 25 μL with the following R-PCR program (42°C, 10 minutes; 95°C, 10 seconds; 95°C 10 seconds, 60°C, 1 minute) for 40 cycles. The primers and probes are described in Table 1.
| Primer | Sequence |
|---|---|
| Genotyping | |
| Sense | GGTGGTTTCTGGGGTGAC |
| Antisense | AGCCGACGAAATYAATTCTGTC |
| R-PCR | |
| Sense | GGTGGTTTCTGGGGTGAC |
| Anti | AGGGGTTGGTTGGATGAA |
| Probe | FAM-TGATTCTCAGCCCTTCGC-MGB |
| F1 | |
| Sense | TAATACGACTCACTATAGGGCAGACCACGTATGTGGTCGAC |
| Anti | TGCACAGGGACGGCATGAAA |
| F2 | |
| Sense | TTTCATGCCGTCCCTGTGCA |
| Anti | CCTCAAGCCGCTTAGGGTTATGTTC |
| F3 | |
| Sense | GAACATAACCCTAAGCGGCTTGAGG |
| Anti | GCAGCCTTCGCCGCCTGAG |
| F4 | |
| Sense | CTCAGGCGGCGAAGGCTGC |
| Anti | CGGATCATCCACTGCGGCAT |
| F5 | |
| Sense | ATGCCGCAGTGGATGATCCG |
| Anti | GTCACCCCAGAAACCACC |
| F6 | |
| Sense | GGTGGTTTCTGGGGTGAC |
| Anti | AGCCGACGAAATYAATTCTGTC |
| F7 | |
| Sense | GCTTCCGACAGAATTRATTTCGTCGGC |
| Anti | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCAGGGAGCGC |
2.4 HEV genotyping
For HEV genotyping, cDNA was used as template for PCR amplification with superFi DNA polymerase (Invitrogen) in a 25 μL reaction mixture as follows: 5 × SuperFi buffer, 5 μL; GC enhancer, 5 μL; 10 mM dNTP mix, 0.5 μL; primer mix (10 μM each) (Table 1), 2.5 μL; template cDNA, 1.5 μL; SuperFi polymerase, 0.5 μL. The primer sequences are as described in Table 1. PCR amplification was carried out by using the program as follows: 98°C 30 seconds; 98°C 10 seconds, 58°C 10 seconds, 72°C 1 minute, 45 cycles; 72°C 10 minutes. The PCR products were gel purified and ligated into a homemade cloning vector pZero-blunt. Colonies were propagated and the plasmid was prepared and sequenced by Sanger sequencing with primers on the vector backbone.
2.5 Cloning and sequencing the full-length HEV genome
For cloning the full-length HEV genome, 2 μL of cDNA was used as a template for PCR amplification with superFi DNA polymerase in a 25 μL reaction mixture, as described above with primers for seven fragments encompassing the entire viral genome (Table 1). PCR amplification was carried out by using the program as follows: 98°C 30 seconds; 98°C 10 seconds, 58°C 10 seconds, 72°C 1 minute, 40 cycles; 72°C 10 minutes. PCR products were gel purified and ligated into pZero-blunt. For each cloned fragment, at least six colonies were propagated and sequenced by Sanger sequencing with primers on the vector backbone. For sequencing certain inserts, additional internal sequencing primers were designed based on the sequenced sequences.
2.6 Bioinformatics
DNA sequences were aligned and the contigs were assembled by Seqman (DNAstar). DNA sequences were analyzed and translated by Editseq (DNAstar). The assembled HEV4-js1 sequence was aligned with HEV sequences in the database (https://www.viprbrc.org) and similar HEV sequences including each representative HEV4 subgenotypes were extracted for the generation of the phylogenetic tree with Mega10 (www.megasoftware.net).
3 RESULTS
3.1 Patient history
A 31-year-old male was admitted to the hepatitis department of a local hospital for the first time in November 2017 because of fatigue and decreased appetite persisting for 1 week. On examination, he had yellow skin mucosa and sclera and yellow urine. The other vital signs were normal. He reported a history of heavy drinking. Liver function tests revealed an acute rise of the liver enzyme alanine aminotransferase (ALT, 1300 U/L) and total bilirubin (130 μmol/L). The antibody test of hepatitis A virus, hepatitis C virus, hepatitis E virus, Treponema pallidum, and HIV were negative, except that for the hepatitis B virus surface antibody was positive and the hepatitis B surface antigen test was negative. Autoantibodies for autoimmune hepatitis were negative. Abdominal ultrasound indicated an enlarged echo of liver parenchyma. Eventually, he was diagnosed with alcoholic hepatitis. After intravenous administration of compound glycyrrhizin and polyene phosphatidylcholine by his physician, the patient's clinical symptoms gradually relieved and he was discharged home with a prescription for oral hepatoprotective drugs and alcohol abstinence.
His liver function returned to be normal in early April 2018. However the levels of his liver enzyme ALT rose again to 260 U/L in the late April 2018. From then on, he suffered from fatigue and discomfort again. This patient was transferred to the hepatitis department of our hospital for further treatment in July 15, 2018. He denied chronic liver disease and familial hereditary liver disease. A thorough survey of food and toxicant consumption such as the consumption of red meat, wild boar, raw or poorly cooked seafood, raw fruit, and vegetables or hepatotoxic substances was conducted, but the results revealed no obvious possibility of transmission through these routes. The patient had no history of blood transfusions and no history of travelling abroad. Physical examination was normal and the liver was not palpable.
3.2 Diagnosis of this patient as an acute HEV infection
On July 19, 2018, detection of anti-HEV IgG and IgM antibodies in the patient's serum showed that both anti-HEV IgG (S/CO, 11.9) and IgM (S/CO, 213.5) were positive. The ALT (257 U/L) was abnormal (Figure 1). Examination of the HEV viral genome in the plasma and feces by reverse transcription and quantitative real-time PCR (RT-rPCR) showed positive viral RNA in the feces with a Ct value of 26.7 whereas negative viral RNA in the plasma sample probably due to short-lived viremia10 (Figure 1). On July 31, 2018, the anti-HEV IgM dropped to 157(S/CO) whereas the anti-HEV IgG increased to 12.6 (S/CO). Meanwhile, the ALT level (24 U/L) dropped to a normal level and there was still positive viral RNA in the feces with a Ct value of 28.9 whereas negative viral RNA in the plasma sample. Considering the recurrence of liver dysfunction in the patient, we did a liver biopsy. The hepatic tissue pathology suggested a fatty degeneration of hepatocytes and intrahepatic cholestasis hepatitis (G2S2). Viral RNAs could be detected in the liver tissue with a Ct value of 27.4 (Figure 1). On August 11, 2018, the anti-HEV IgM (S/CO, 137.0) further dropped and the anti-IgG (S/CO, 14.5) further increased. The ALT level (13 U/L) remained at a normal level. The detection of the viral genome in the plasma and feces was negative. The patient was discharged home. On November 7, 2018, the patient revisited the hospital. The anti-HEV IgM (S/CO, 30.1) dramatically dropped and the anti-HEV IgG (S/CO, 19.7) further increased. The fecal samples were detected for viral RNA and the result was negative. Collectively, this patient got an acute HEV infection and recovered from the acute infection.
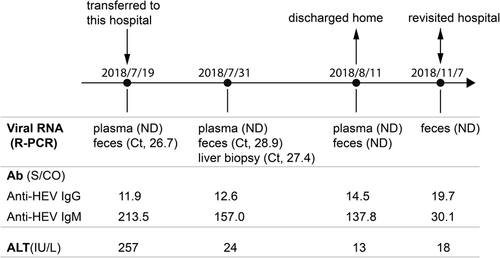
3.3 Cloning the entire genome of HEV and phylogenetic analysis of the HEV
We first carried out the genotyping of the HEV. RNAs from the fecal sample were reversely transcribed to cDNA. An HEV fragment was amplified with two conserved primers (Table 1). The amplified fragment was cloned and sequenced. Sequence alignment showed the sequence was highly similar to sequences of HEV genotype 4.
Based on the sequences of HEV genotype 4 in the database, we designed primers to amplify the entire HEV4 genomes (Figure 2). We extracted total RNAs from the feces sample during the icteric phase when the ALT was at a maximum plateau and the liver biopsy sample during the convalescent phase when the ALT level dropped back to normal and anti-HEV IgG began to increase (Figure 1). Extracted RNAs were reversely transcribed with random primers. Seven fragments encompassing the entire HEV genome were successfully amplified from the cDNA with the designed primers (Table 1) and each fragment was cloned. We failed to amplify viral fragments from the fecal samples during the convalescent phase, probably due to technical issues (Figure 1). To capture the potential quasispecies of HEV, for each cloned amplicon, we sequenced multiple colonies by Sanger sequencing. The assembled consensus sequence was named HEV4-js1.
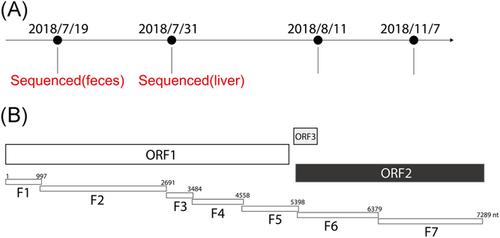
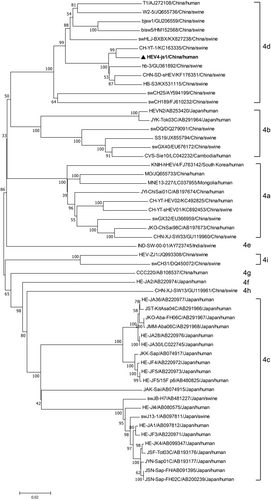
Then, we generated the phylogenetic tree of the full-length nucleotide sequence of the HEV4-js1 with the reported HEV4 sequences. The HEV4-js1 was most related to an HEV4 CH-YT-HEV01 (subgenotype 4d), isolated from humans in Shangdong Province of China11 (Figure 3). The CH-YT-HEV01 strain is very close to a swine isolate HB-3 (GU361892), which was first found from swine in Hubei Province of China.11 The HEV4-js1 was also very close to the HB-3 (Figure 3), suggesting a potential trans-regional transmission and cross-species transmission.
3.4 HEV quasispecies dynamics during the acute HEV infection
There is evidence for the presence of HEV quasispecies that may favor virus-host interaction.8, 9, 12 By sequencing multiple colonies of each amplified HEV fragment, we found genetic variations that change the amino acids in the proline-rich domain (PCP), hypervariable region (HPX) and RNA-dependent RNA polymerase (RdRp) region in the ORF1 and in the P domain in ORF2 either from the feces or from the liver biopsy (Figure 4A,B). During the icteric phase, in the fecal sample, there were only two amino acid variations (A557T, L717P) in the PCP and HPX regions in ORF1 and four amino acid variations (S582F, A606V, F626L, A645V) in the P domain in ORF2. In contrast, during the convalescent phase, in the liver biopsy sample, there were five amino acid variations (V659A, R703Q, V766A, P770S, F854S) in the HPX region and three amino acid variations (L1352F, L1557I, I1681V) in the RdRp region in ORF1 and two amino acid variations (K518Q, D587N) in the P domain in ORF2 (Figure 4A,B). These results suggest broader genetic variations in the ORF1 region during the convalescent phase whereas broader genetic variations in the ORF2 region during the icteric phase.
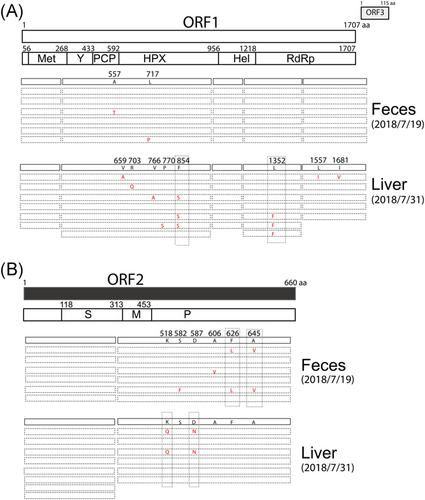
3.5 Hot spot genetic variation sites in ORF1 and ORF2
Of these amino acid variations, there were dominant variations found in at least two sequenced colonies, such as the F854S in the HPX region and L1352F in the RdRp region in ORF1 and K518Q, D587N, F626L and A645V in the P domain in ORF2 (Figure 4A,B).
HEV ORF2 consists of the S domain (aa 118-313), M domain (aa 314-453) and the P domain (aa 454-606) that forms the protrusion projecting from the viral shell.1 The P domain contains numerous neutralizing sites.13 Two dominant amino acid variations F626L and A645V found in the fecal samples during the icteric phase and two dominant amino acid variations K518Q, D587N found in the liver biopsy sample during the convalescent phase are located in the P domain (Figure 4B).
4 DISCUSSION
It has been reported that the dynamics of the viral quasispecies predict the outcome of acute hepatitis C.14 High quasispecies heterogeneity is associated with persistent HEV infection.12 HEV in chronically infected patients has higher complexity and diversity than HEV in the acutely infected patients.9 However, the HEV quasispecies dynamics during an acute HEV infection is poorly understood.
In this study, we found the existence of HEV quasispecies and different quasispecies patterns during the icteric phase and convalescent phase. There were broader genetic variations (eight amino acid variations) in the ORF1 region during the convalescent phase than that (two amino acid variations) during the icteric phase (Figure 4A). Unfortunately, we failed to clone the viral genome from the fecal sample during the convalescent phase and could not directly compare the sequences of the virus in feces and liver during the convalescent phase. Once released from infected hepatocytes into the bile canaliculi, HEV is released through the intestine and excreted into stools. Very recently it was reported that HEV replicates in human intestinal cells in vitro.15 The virus in the feces is either released from hepatocyte or from intestinal cells. We noticed that there were no overlapped genetic variations between the feces and the liver biopsy samples (Figure 4). It is possible that virus released from the intestinal into the feces is different from that released from hepatocyte and these two virus population may have diffident genetic variation pattern.
The broader genetic variations in the nonstructural protein ORF1 region during the convalescent phase might imply less T-cell mediated immune pressure during the convalescent phase. However, during the icteric phase of hepatitis E, CD8+ T cells may recognize viral variants and respond efficiently to these epitopes as in HIV infection.6 In HIV-infected patients, escape mutations in the viral Gag protein dramatically reduce viral replication capacity.16, 17 Whether or not these genetic variations observed in the ORF1 influence the viral replication needs further studies.
In contrast to ORF1, during the icteric phase, there were broader amino acid variations (S582F, A606V, F626L, A645V) than that during the convalescent phase (K518Q, D587N) in the P domain in ORF2 (Figure 4B), suggesting strong B-cell-mediated evolution of viral surface epitopes associated with viral clearance in vivo, as the P domain contains numerous neutralizing sites.13
In summary, we studied the HEV quasispecies dynamics during acute infection and uncovered different quasispecies patterns during the icteric phase and convalescent phase, which is analogous to persistent HEV infection. The quasispecies dynamics might reflect host immune pressure during the viral clearance.
ACKNOWLEDGMENTS
This study was supported in part by the National Megaprojects of China for Infectious Diseases (grant number 2017ZX10103009 to YZ). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceived the study: ZY; conducted the study: FD, XM, JZ, ZY; data analysis: XM, FD, ZY; manuscript draft: XM, ZY; resources: ZQ, ZY, ZY.