Measles virus genotypes circulating in India, 2011–2015
Abstract
The Government of India is accepted to participate in the measles elimination and rubella control goal 2020, hence genetic characterization of measles viruses (MeV) becomes essential. At National Reference Laboratory (National Institute of Virology, Pune), the throat swabs/urine specimens (n = 380) or PCR products (n = 219) obtained from the suspected measles cases were referred for the molecular testing and subsequently, MeV nucleoprotein (N) gene sequencing/genotyping. In addition, 2,449 suspected measles cases, mainly from the Maharashtra state were referred for the laboratory diagnosis. A detailed study was performed on N gene sequences obtained during last two decades. Indian MeV sequences obtained during 2011–2015 were compared with 1996–2010 sequences and genetic divergence was studied. Circulation of measles genotypes B3 (n = 3), D4 (n = 49), and D8 (n = 351) strains were observed in 19 States and three Union Territories of India. In addition, 64 measles viruses were isolated from 253 throat swab or urine specimens obtained from the suspected measles cases. During 2011–2015, 67.9% (1,663/2,449) suspected measles cases were laboratory confirmed. Molecular studies showed circulation of measles genotype B3 in India along with prominently circulating genotypes D4 and D8 except D7 strains. The genetic diversion within Indian B3, D4, and D8 genotypes was 0.3%, 1.1%, and 2.1%, respectively. The genetic divergence of Indian B3, D4, and D8 measles strains with the WHO reference sequences was 2.5%, 2.6%, and 1.8%, respectively. It is crucial data for national immunization program. More measles/rubella genotyping studies are necessary to track transmission and to support measles elimination and rubella control. J. Med. Virol. 89:753–758, 2017. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Measles virus (MeV) is highly infectious and causes high grade fever, cough, coryza, conjunctivitis, and maculopapular rashes in humans. MeV belongs to the family Paramyxoviridae and genus Morbillivirus. It is enveloped, single stranded, negative sense RNA virus with 15.8 Kb genome size. Measles is serologically monotypic virus but distributed into eight clades (A–H) and 24 subclades or genotypes [WHO, 2012, 2015]. Measles is an endemic disease in India causing significant morbidity and mortality; hence, Government of India (GoI) is working for measles elimination and rubella control in the phased manner [Vaidya, 2015]. Recently, GoI had introduced two doses of measles containing vaccine in Universal Immunization Program (UIP). Similarly, measles case-based surveillance is being initiated in different parts of the country. For the genetic characterization of measles viruses, clinical specimens and PCR products were referred at the World Health Organization accredited National Reference Laboratory (NRL) at National Institute of Virology (NIV), Pune.
Molecular epidemiology of measles in India has been considerable growing and many measles virus sequences are available in the GenBank [Wairagkar et al., 2011, 2002]. Phylogenetic studies revealed circulation of measles genotypes D4 and D8 strains. Measles genotype D7 was detected from Bengaluru, Chennai, and Pune cities [Vaidya et al., 2008]; however, circulation of D7 genotype was not detected afterwards. Molecular studies from the States of Uttar Pradesh [Shakya et al., 2012a,2012b] and Tamil Nadu [Ramamurty et al., 2006; Duraisamy et al., 2012] showed presence of measles genotype D8 strains. India's MeV sequencing data now covers different regions of the country. Recently, circulation of measles genotype B3 was reported from Thiruvananthapuram, Kerala [Kuttiatt et al., 2014] indicating either importation from other countries or unidentified indigenous measles strain.
The World Health Organization South-East Asia Region has committed to eliminate measles by 2020. The WHO Global Measles and Rubella Laboratory Network (GMRLN) was established to provide high quality laboratory support for surveillance [Featherstone et al., 2003; Perry et al., 2015; Mulders et al., 2016]. Similarly, the GoI is working for measles elimination and rubella control goal. GoI had introduced one dose of measles vaccine at the age of 9 months in its UIP during 1985. Second dose of measles was introduced in 2010, primarily in the states where measles vaccine coverage is <80% and subsequently in other States. Recently, the GoI announced to introduce the rubella vaccine in its UIP [Press Information Bureau Government of India, 2014]. As per the revised immunization schedule, first dose was offered at the age of 9–12 months and second dose at the age of 16–24 months. If a child has missed the first and second dose, both doses can be offered up to 5 years of age maintaining a gap of at least 4 weeks between the doses. The Indian Academy of Pediatrics (IAP) supports the elimination of measles and rubella but also of mumps in the form of two doses of affordable measles-mumps-rubella (MMR) vaccine [Vashishtha et al., 2014].
As a part of genetic characterization of measles viruses, the clinical specimens or PCR products were referred to NRL for the N gene sequencing and genotyping. MeV sequences obtained during 2011–2015 were compared with previously published sequences including WHO standard reference strains and named strains available in the MeaNS database (www.who-measles.org) [Rota et al., 2011]. A detailed study was undertaken on measles N gene sequences detected from India during last two decades.
MATERIALS AND METHODS
Serum Samples Referred for Laboratory Diagnosis
As a part of WHO, India supported measles outbreak surveillance, suspected measles outbreaks are investigated and clinical specimens were sent to the nearest network laboratory. Measles reporting network includes, Government health facilities, Private health facilities, Indian Systems of Medicine practitioners, religious and traditional healers, and others. Laboratory network consists of two National Reference Laboratories (Kings Institute of Preventive Medicine and Research, Chennai for serology and National Institute of Virology, Pune for molecular testing) and eleven National Laboratories situated across the country. These laboratories primarily helps in the serological (measles/rubella IgM EIA) testing. NIV Pune is providing serological testing for Maharashtra state and also providing support for the molecular testing (i.e., measles/rubella virus genotyping and sequencing) to entire country.
For laboratory investigation, serum samples were collected after obtaining informed consent from patients or from their parents in case of minors. Serum samples were collected from the suspected measles cases (i.e., fever with rash and cough/cold/coryza/conjunctivitis) and representative samples (n = 3–5) from each outbreak were transported within 12 hr on ice packs to the NRL. All the suspected serum samples were subjected to measles virus IgM antibody detection (Siemens Healthcare Diagnostics Products GmbH, Germany). Measles negative and equivocal samples were tested for the rubella specific IgM antibody. Majority of the serum samples were referred from different districts of Maharashtra state.
Measles RT-PCR and Sequencing
Throat swabs or urine specimens were referred from the suspected measles cases (n = 380) and PCR products (n = 219) received from the National Measles Laboratories [NIV Unit Bengaluru, n = 66; KIPM&R Chennai, n = 78; SGPGIMS Lucknow, n = 72; Government Medical College Guwahati, n = 3] were subjected to measles virus sequencing and genotyping using the WHO standard protocols [WHO, 2015]. Briefly, viral RNA was extracted from 140 μl of throat swab or urine sample according to the manufacturer's instructions (Qiagen, Hilden, Germany). RNA was reconstituted in 60 μl nuclease free water and stored at −70°C until tested. Measles virus N gene was amplified using primers; MeV216-5′ TGG AGC TAT GCC ATG GGA GT 3′ and MeV214-5′ TAA CAA TGA TGG AGG GTA GG 3′. One step RT-PCR was performed using Super-Script III One-Step RT-PCR System with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). The complementary-DNA was prepared at 55°C for 30 min followed by hot-start of the PCR cocktail at 94°C for 2 min and 40 cycles of 94°C for 15 sec (denaturation), 55°C for 30 sec (annealing), 72°C for 30 sec (extension), and a final extension step at 72°C for 7 min was completed in a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). The PCR products were purified using the QIAamp DNA minikit and sequenced in both directions using same primer pair using reaction conditions; 25 cycles of 94°C for 10 sec, 50°C for 5 sec and 60°C for 4 min. The sequencing reaction was performed using the DyeDeoxy terminator sequencing kit (Applied Biosystems). Furthermore, a sequencing reaction (forward and reverse) was purified using a Dye Ex 2.0 Spin Kit. The N gene sequences were obtained using an automated sequencer (ABI/HITACHI 3730XL, DNA Analyzer (96), Tokyo, Japan). The consensus sequences were subjected to phylogenetic analysis using the WHO standard reference sequences.
Serum samples were not used for the measles virus RT-PCR sequencing or genotyping studies. Throat swabs or urine specimens were referred from 19 States and three Union Territories of India. A suspected case detail was not available for the PCR products referred by the National laboratories.
A phylogenetic tree of N gene was constructed by using the neighbor-joining and Kimura two-parameter methods. The robustness of the grouping was assessed by using 1,000 bootstrapping replicates.
Measles Virus Isolation
A total of 253 of 380 throat swab or urine specimens were used for the virus isolation, that is, 3 from 2011, 9 from 2012, 26 from 2013, 136 from 2014, and 79 from 2015. Five hundred microliter of processed throat swab or 1 ml of centrifuged urine specimen was inoculated into the flask(s) containing 24 hr old Vero hSLAM cell monolayer and cytopathic effect (CPE) was observed up to 6 days. Specimens with negative CPE were passage three times to confirm virus isolation. The CPE positive cell suspensions were confirmed by measles RT-PCR and sequencing.
RESULTS
Epidemiological Information
Between 2011 and 2015, serum samples from suspected measles cases were received at NIV Pune for the serological testing. Majority of suspected cases were from Maharashtra state. Altogether, 2,449 serum samples were obtained from the suspected measles cases that consist of 1,180 females and 1,269 males aged between 8 months and 25 years. The frequency of samples received for the laboratory diagnosis was highest (34%) in year 2014, followed by 26.7% in 2011 and lowest (3.4%) in year 2012 (Table I). Measles outbreak based or case based surveillance was not in the place during 2012, due to which serum sample number was limited (only sporadic cases referred from the local hospitals were tested). Of these suspected cases, 626 had history of at least one dose of measles vaccine as per the record or by the parents recall; however, measles vaccination history was either unknown or not available for the 1,823 cases. During this period, 599 clinical specimens (i.e., throat swabs or urine specimens) collected from the suspected measles cases and PCR products were referred for measles virus detection by RT-PCR, genotyping/sequencing, and virus isolation. These clinical samples were received from 19 States and three Union Territories of India. The frequency of clinical specimens received for the virus detection/isolation and genotyping was higher in the year 2014 and 2015 (79.1%, 476/599) due to special efforts by the field epidemiological team (NPSP project, WHO India). During 2011–13, 123 clinical specimens were referred for virus isolation and genotyping (Table II).
| Year | Suspected cases | Lab confirmed measles cases | Lab confirmed rubella cases | Non-measles/non-rubella cases |
|---|---|---|---|---|
| 2011 | 654 | 482 | 44/178 | 128 |
| 2012 | 83 | 48 | 6/35 | 29 |
| 2013 | 534 | 355 | 74/181 | 105 |
| 2014 | 834 | 557 | 37/286 | 240 |
| 2015 | 344 | 221 | 44/340 | 79 |
| Total | 2449 | 1663 | 205/1020 | 581 |
| Year | Clinically suspected cases tested by RT-PCR | PCR positive specimens/PCR products | MeV genotypes detected |
|---|---|---|---|
| 2011 | 46 | 13 | 7 D8 and 6 D4 |
| 2012 | 38 | 13 | 8 D8 and 5 D4 |
| 2013 | 39 | 19 | 10 D8 and 9 D4 |
| 2014 | 180 | 119 | 108 D8, 10 D4, and 1B3 |
| 2015 | 296 | 239 | 218 D8, 19 D4, and 2B3 |
| Total | 599 | 403 | 351 D8, 49 D4, and 3B3 |
Laboratory Confirmed Measles Cases
During 2011–2015, 67.9% (1,663/2,449) suspected measles cases were laboratory confirmed, 20% (205/1,020) cases showed rubella infection, and 23.7% suspected cases could not confirmed for both measles and rubella (Table I). Age wise distribution of measles cases indicated higher percentage in 1–14 years (86.9%, 1,445/1,663) and lower in 0–11 months (8.1%, 135/1,663) and 15+ years (5%, 83/1,663). Among the immunized individuals (n = 626), 385 had laboratory confirmed measles infection and majority of these cases (n = 367) were 1–14 years old.
MeV Isolates Obtained During 2011–2015
Altogether, 253 throat swab or urine specimens were cultured in 24 hr old Vero hSLAM cells and 64 measles viruses were isolated. Nine measles isolates were obtained in year 2012, 5 isolates in 2013, 32 isolates in 2014, 18 isolates in 2015, and none from 2011. Twenty-five isolates were obtained from Maharashtra, 13 from Gujarat, 7 from Madhya Pradesh, 5 from Karnataka, 3 each from Uttarakhand, Chhattisgarh, and Dadra and Nagar Haveli, and 1 each from Odisha, Delhi, Haryana, Rajasthan, and Daman and Diu. Total of 14 MeV genotype D4 isolates and 50 MeV genotype D8 isolates were obtained during this period. History of measles vaccination (by documentation or parent's recall) was available for 14 individuals, of which two were D4 isolates and ten were D8 isolates.
Detection of MeV Genotypes
During 2011–2015, about 12 measles genotype B3 strains were detected from three different States. Two measles strains were detected from the active surveillance conducted in Andhra Pradesh and Uttar Pradesh (Fig. 1). Previously, ten measles B3 strains were detected in the archived samples from Kerala [Kuttiatt et al., 2014]. Comparison of these B3 sequences with two WHO reference sequences and seven named strains available in the MeaNS [Rota et al., 2011] revealed close identity with MVs/Tonbridge.GBR/5.14, MVi/Harare.ZWE/38.09, MVs/Kansas.USA/1.12, and MVs/Western Australia.AUS/2.14 strains (Fig. 2). The genetic divergence (P-distance) within Indian B3 strains was 0.003 and between the WHO reference strains and Indian B3 strains was 0.025.
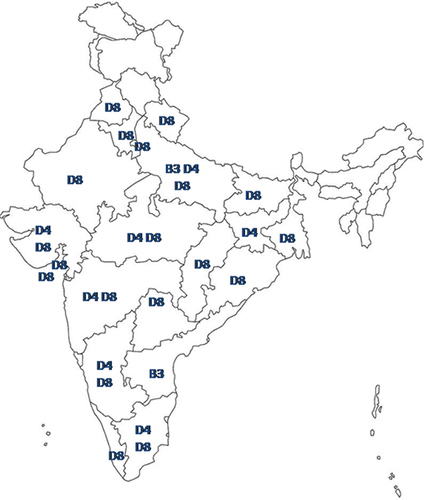

Altogether, 49 measles genotype D4 strains were detected from the States of Gujarat, Jharkhand, Karnataka, Madhya Pradesh, Maharashtra, Rajasthan, Tamil Nadu, and Uttar Pradesh (Fig. 1). Majority of the measles D4 strains (n = 44) were detected from the States of Gujarat, Karnataka, and Maharashtra. Comparison of these D4 sequences with the WHO reference sequence and ten named strains available in the MeaNS [Rota et al., 2011] revealed close identity with MVi/Montreal.CAN/0.89, MVs/Birmingham.GBR/7.09, and MVs/Madrid.ESP/46.10 strains (Fig. 2). The genetic divergence (P-distance) within Indian D4 strains was 0.011 and between the WHO reference strains and Indian D4 strains was 0.026.
Circulation of measles genotype D8 strains were detected from 20 States and Union Territories except Andhra Pradesh and Jharkhand states (Fig. 1 and Table II). Majority of measles D8 strains were detected from the nine States (i.e., Bihar, Delhi, Gujarat, Karnataka, Maharashtra, Madhya Pradesh, Odisha, Tamil Nadu, and Uttar Pradesh) with overall contribution of 88.3% (310/351). Comparison of these D8 sequences with the WHO reference sequence and eleven named strains available in the MeaNS [Rota et al., 2011] revealed close identity with MVi/Manchester.GBR/30.94, MVs/Victoria.AUS/6.11, MVs/Pernambuco.BRA/25.13/6, MVs/Chui.KGZ/53.14, and MVs/Frankfurt Main.DEU/17.11 strains. The genetic divergence (P-distance) within Indian D8 strains was 0.021 and between the WHO reference strains and Indian D8 strains was 0.018.
DISCUSSION
In India, first dose of measles vaccine has been provided at the age of 9 months through UIP in 29 States and seven Union Territories. In 2010, following National Technical Advisory Group on Immunization (NTAGI) recommendations, the GoI introduced two dose measles strategy in phased manner. Measles is serologically monotypic virus, thus available vaccines can protect other circulating wild types. However, measles infection in the vaccinated population may be due to; primary vaccine failure, secondary vaccine failure (waning of immunity), or other unknown reasons. In present study, individuals with a history of measles vaccination during childhood (by documentation or parents recall) showed laboratory confirmed measles infection. However, proper information about the number of measles vaccine dose(s) was not available. It is important to note that measles vaccine was introduced in India during 1985 in the form of one dose of measles at the age of 9 months. As per the WHO recommendation, at least two doses of measles vaccine should be offered to maintain the sufficient immunity levels or protective titer [Thapa et al., 2015]. The WHO developed a measles programmatic risk assessment tool to support measles elimination efforts, and this tool was pilot tested in the state of Uttarakhand, India [Goel et al., 2016].
The genetic characterization of wild type measles virus is an important tool for establishing epidemic links, tracking transmission pathways, understanding importation or exportation of cases, and identifying cases associated with vaccine or wild type viruses [WHO, 2015]. Presently, measles outbreak based surveillance has been conducted in different parts of the country by utilizing local surveillance system or existing polio surveillance program [Shaikh et al., 2015; Vaidya, 2015; Vaidya et al., 2016a,2016b]. The GoI is planning to launch measles case based surveillance in the country. It may provide a true picture of measles in the country with line listed cases coupled with adequate laboratory support. Case-based measles surveillance was performed during November 2009 to December 2011 in the Pune city (India), may provide some useful information before entering into the measles elimination [Bose et al., 2014]. It is believed that GoI may start case-based surveillance at the end of 2016.
During 1996–2010, 196 D8 and 89 D4 strains were detected from 19 States and UT's [Wairagkar et al., 2011]. In addition, circulation of three measles genotype A [Wairagkar et al., 2002; Shakya et al., 2012a] and five D7 strains were detected from Maharashtra, Karnataka, Tamil Nadu, and Uttar Pradesh. During this study (2011–2015), circulation of measles genotype B3 (n = 3), D4 (n = 49), and D8 (n = 351) were detected from 22 States and UT's (Table II). However, limited epidemiological data are available from these States and a scale of outbreaks was not known. Interestingly, measles genotyping data have been added from six States (i.e., Haryana, Jharkhand, Punjab, Rajasthan, Telangana, and Uttarakhand) and three Union Territories (i.e., Delhi, Dadara and Nagar Haveli, and Daman and Diu). Altogether, 661 MeV sequences were detected from different parts of the country during 1996–2015 [GenBank Database http://www.ncbi.nlm.nih.gov/]. Three measles genotype A, eleven B3, 138 D4, seven D7, and 504 D8 sequences are deposited in the GenBank/MeaNS database. For the genetic and antigenic characterization of measles wild types, virus isolation plays a key role. Thus, virus stocks may be useful for studying circulating viruses in pre- and post-elimination era. The complete genome sequence of the circulating viruses may be useful for new developments in the field of vaccine and antiviral discovery.
Previously, circulation of measles B3 strain was detected from the state of Kerala [Kuttiatt et al., 2014] and recently, from Uttar Pradesh and Andhra Pradesh states. Globally, circulation of measles genotype B3 has been reported from many countries, so difficult to understand the indigenous nature of virus or its importation from other countries due to tourism or movement due to employment. However, Indian measles B3 strains had low diversity (0.3%) among each other, whereas 2.5% diversity with the WHO standard reference sequences detected from Zimbabwe, UK, USA, and Australia. Since long time, circulation of measles genotypes D4 and D8 has been reported from India with divergence of 1.1% in Indian D4 strains and 2.1% in Indian D8 strains. Thus, genetic characterization of measles strains would strengthen molecular epidemiology of measles and may provide more information about the importation or exportation of wild type viruses.
In conclusion, circulation of three measles genotypes (B3, D4, and D8) was detected in last 5 years from India. However, case based measles surveillance may show a true picture of currently circulating measles viruses in India. Thus, baseline data on circulating wild type measles or rubella viruses are essential for strengthening national immunization program and ultimately useful for measles elimination and rubella goal 2020.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Sirima Pattamadilok (WHO SEARO) and Dr. Lucky Sangal (WHO India) for their support during measles surveillance program. The authors acknowledge laboratory support of Mr. Madhukar Kamble and Mrs. Neelakshi Kumbhar and bioinformatics support of Mr. Santoshkumar Jadhav.




