Phylogenetic analysis of the NS5 gene of Zika virus
Abstract
ZIKV infection has become a global threat spreading across 31 countries in Central America, South America, and the Caribbean. However, little information is available about the molecular epidemiology of ZIKV. Shared mutation of a threonine residue to alanine at the same position in the C terminal of NS5 sequences was observed in sequences from Colombia, Mexico, Panama, and Martinique. The sequences in the phylogenetic tree fell within the same cluster. Based on shared mutation the presence of a Latin American genotype was proposed. Comparison of African and Asian lineages yielded R29N, N273S, H383Q, and P391S mutation. The study highlights that mutation of amino acids at NS5 may contribute to neutropism of ZIKV. J. Med. Virol. 88:1821–1826, 2016. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Since its appearance in humans in 1954 [Macnamara, 1954], ZIKV infection has become a global health threat. It has spread outside Africa and Asia and is no longer confined to these regions [Haddow et al., 2012; Faye et al., 2014]. The first pacific outbreak was reported in Yap island in the Federated states of Micronesia in 2007 [Duffy et al., 2009] and in French Polynesia in 2013 [Musso et al., 2014; Cao-Lormeau et al., 2016; Malkki, 2016]. It had spread to New Caledonia, Cook island, and Easter island [Musso et al., 2015] and later appeared in Brazil [Campos et al., 2015; Zanluca et al., 2015]. However, the Brazilian outbreak (2015–2016) turned pandemic with autochthonous transmission of ZIKV causing microcephaly in newborns [Mlakar et al., 2016]. ZIKV has been associated with a rapidly developing neurotropic threat causing microcephaly and Guillain–Barré syndrome [Calvet et al., 2016; Cao-Lormeau et al., 2016]. It spread across Latin American countries of Colombia, El Salvador, Suriname, and Martinique (WHO situation report, March 17, 2016). Acute illness and myelitis due to ZIKV have been reported from Latin American countries and the Carribean [Araúz et al., 2016; Rodriguez-Morales et al., 2016; Rozé et al., 2016] and a global public health emergency was declared in Feb, 2016 [WHO, 2016].
With the onset of severe forms of ZIKV, it was decided to compare NS5 protein sequences of ZIKV isolates from different countries. The ZIKV NS5 gene contain unique phylogenetic signals and were found suitable for analysis [Faye et al., 2014].
METHODS
The NS5 full length sequences were 652 amino acid in length. The partial NS5 sequences were aligned at amino acids 1–466 of the full NS5 gene. All protein sequences were aligned using MUSCLE under default settings. The evolutionary history was inferred by using the Maximum Likelihood method based on the Le_Gascuel_2008 model. Evolutionary analyses were conducted in MEGA6. The analysis involved 55 sequences of ZIKV from GenBank. The tree was presented using Figtree.
RESULTS AND DISCUSSION
Phylogenetic trees identified two major lineages, the Asian and the African lineage (Fig. 1A) consistent with previous reports [Haddow et al., 2012; Faye et al., 2014]. The present study was built up from previous analysis of Faye et al. [2014]; Wang et al. [2016]; and Zhang et al. [] (Fig. 1A) using additional sequences updated up to May 2016. The sequences from Colombia, Mexico, Panama, and Martinique (Latin American countries) formed a separate clade distinct from Brazilian sequences (Fig. 1A) referred as Latin American strain. Recent ZIKV infection reported in Central America and the French West Indies especially Martinique have been associated with neurological complications and acute symptoms associated with Guillain–Barré syndrome [Araúz et al., 2016; Rozé et al., 2016; WHO situation report, March 2016]. The amino acid polymorphism exhibited by NS5 of ZIKV in the alignment showed T581A substitution in nine sequences obtained from isolates of Colombia, Martinique, Mexico, and Panama (Fig. 1B). The substitution lie in the C-terminal of NS5 gene involved in RNA-dependent RNA polymerase activity. The domains of the enzyme in flaviviruses are involved in intermolecular interaction and is responsible for de novo RNA synthesis of flaviruses [Zou et al., 2011]. It is suggested that mutation in the C-terminal of NS5 may bring about changes in RNA replication since hydrophilic to hydrophobic substitution was observed. The sequences in the Latin American genotype which are of clinical origin include AMQ34004, AMQ34003, AMM39804, AMZ03557, AMC33116, ANB66184, ANB66182, ANB66183. Although Zhang et al. [] did similar work of clustering strains from Colombia, Mexico, and Martinique, they failed to observe the amino acid mutation T581A, since they did not use the protein sequence of NS5 in the analysis. Their group also did not include sequences from Panama isolates which also bears T581A mutation (Fig. 1B). The polymorphism at the amino acid level was useful in genotyping the strains as Latin American strains. However, some sequences which clustered together with Brazilian sequences and not with the Latin American cluster were the sequences from Mexico (ANF04750), Honduras (ANG09399), Suriname (ALX35659), Colombia (ANF04752), Puerto Rico (AMZ03556), French Polynesia (AHZ13508) (Fig. 1A). The results suggested two genotypes circulating in Colombia and Mexico based on the sequences found in different clusters.
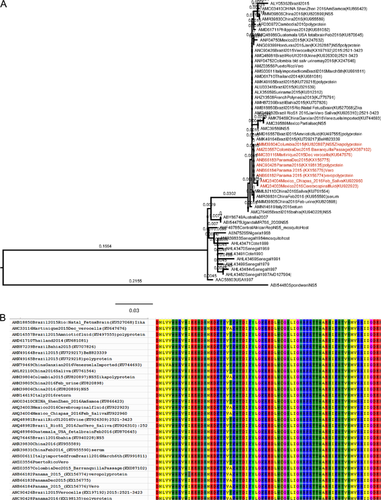
Quiñonez et al. [2016] reported the detection of ZIKV in Mexico from a traveller to Colombia in 2015. The present study also supports this hypothesis that transmission and spread of ZIKV in Mexico may be from the route of Colombia. The sequences from Mexico (AMQ34003 and AMQ34004) belong to the cluster of T581A (Fig. 1A). However, additional amino acid substitution at different positions in the Mexican sequence (AMQ34003 and AMQ34004) revealed H38Q and G359V substitutions occurring at position 38 and 359, respectively (Table I).
| Mutation at amino acid position number indicated | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. | Country/year | Origin country | Clinical source | 29 | 36 | 38 | 60 | 62 | 125 | 138 | 203 | 274 | 359 | 391 | 452 | 581 | |
| 1 | AMK79469 | China, 2016 | Venezuela Imported into China | Serum | N | A | K | D | A | S | H | I | R | G | S | D | T |
| 2 | ALY05362 | Brazil, 2015 | Serum | N | S | H | E | P | S | H | N | – | G | S | D | T | |
| 3 | AMM39806 | China, 2016 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 4 | AMO03410 | China, 2016 | American Samoa, imported into China | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T |
| 5 | AMM39804 | Columbia, 2015 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | ||
| 6 | AAC58803* | USA,1997 | Serum | N | S | H | E | – | – | H | N | R | G | S | D | T | |
| 7 | ABY86749* | Australia, 2007 | Serum | R | S | H | E | P | N | R | N | – | G | – | S | ||
| 8 | AHL43499* | Senegal, 1979 | Serum | N | – | H | – | – | – | R | G | P | S | T | |||
| 9 | AMR39830 | China, 2016 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 10 | AMD61710 | Thailand, 2014 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 11 | AMD61711 | Philippines, 2012 | Serum | N | S | H | E | P | S | H | N | R | G | P | D | T | |
| 12 | ABI54475 | Uganda MR766, 2006 | Serum | R | S | H | E | P | N | R | N | R | G | P | S | T | |
| 13 | AHL43471* | Cote, 1999 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 14 | AHL43470* | Senegal, 1969 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 15 | AHL43469* | Senegal, 1991 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 16 | AHL43484* | Senegal, 1997 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 17 | AHL43482* | Senegal, 1997 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 18 | AHL43491* | Cote, 1990 | Serum | N | – | H | – | – | – | – | – | R | G | P | S | T | |
| 19 | AMB18850 | Brazil, 2015 | Fetus brain | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 20 | AMD16557 | Brazil, 2015 | Amniotic fluid | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 21 | AMK49165 | Brazil, 2015 | Serum | D | S | H | E | P | S | H | N | R | G | P | D | T | |
| 22 | ALX35659 | Suriname, 2015 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 23 | AHZ13508 | French Polynesia, 2013 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 24 | AEN75265 | Nigeria, 1968 | Serum | R | N | H | E | P | N | R | N | R | G | P | S | T | |
| 25 | AMN14619 | Italy, 2016 | Dominican Republic | Urine | N | S | H | E | P | S | H | N | R | G | S | D | T |
| 26 | AMK49164 | Brazil, 2015 BeH823339 | Serum | D | S | H | E | P | S | H | N | R | G | S | D | T | |
| 27 | AML82110 | China, 2016 | Saliva | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 28 | AMD16557 | Brazil, 2015 | Amniotic fluid | N | S | H | V | P | S | H | N | R | G | S | D | T | |
| 29 | AMH87239 | Brazil, Bahia, 2015 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 30 | AMQ34003 | Mexico, 2016 | Cerebrospinal fluid | D | S | Q | E | P | S | H | N | R | V | S | D | A | |
| 31 | AHF49785 | Central African Republic, 2013 | Mosquito host | K | S | H | E | P | N | R | N | R | G | P | S | T | |
| 32 | ACD75819 | Micronesia, 2007 | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 33 | AMQ48981 | Brazil, 2016 | Urine | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 34 | ABI54480 | Spondweni, 2006 | Serum | S | S | Y | E | P | A | N | N | R | G | P | S | T | |
| 35 | AMQ48986 | Guatemala | USA | Fetal brain | N | S | H | E | P | S | H | N | C | G | S | D | T |
| 36 | ANG09399 | Honduras | N | S | H | E | P | S | H | N | C | G | S | D | T | ||
| 37 | ANF04750 | Mexico | N | S | H | E | P | S | H | N | C | G | S | D | T | ||
| 38 | ANF04752 | Colombia | Blood | N | S | H | E | P | S | H | N | R | G | S | D | A | |
| 39 | AFD30972 | Cambodia | Vero cells | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 40 | AMZ03556 | Puerto Rico | Vero cells | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 41 | AMZ03557 | Colombia Barranquilla | Vero cells | N | S | H | E | P | S | H | N | R | G | S | D | A | |
| 42 | AMS00611 | Italy | – | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 43 | AMQ34004 | Mexico | Saliva | N | S | Q | E | P | S | H | N | R | G | S | D | A | |
| 44 | AMM39805 | China | Urine | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 45 | ANB66182 | Panama | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
| 46 | ANB66184 | Panama | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
| 47 | ANC90428 | Panama | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
| 48 | ANB66183 | Panama | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
| 49 | AMC33116 | Martinique | Vero cells | N | S | H | E | P | S | H | N | R | G | S | D | A | |
| 50 | AMR39831 | China | Serum | N | S | H | E | P | S | H | N | R | G | S | D | T | |
| 51 | ANC90426 | Brazil | N | S | H | E | P | S | H | N | R | G | S | D | T | ||
| 52 | AME17085 | Brazil | N | S | H | E | P | S | H | N | R | G | S | D | T | ||
| 53 | AMQ48982 | Brazil | N | S | H | E | P | S | H | N | R | G | S | D | T | ||
| 54 | AMZ03557 | Colombia | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
| 55 | AMC39589 | Mexico | N | S | H | E | P | S | H | N | R | G | S | D | A | ||
- The partial sequences are shown with * and the − dash indicates gaps in the alignment of partial sequences. Origin/country indicated only where travel importation was reported.
Other clusters obtained from the analysis include the sequences from China. ZIKV was introduced into China through more than one route and were important contributory factors for the 2016 spread in China [Zhang et al., ]. Seven sequences from China associated with ZIKV cases were used in the analysis. The present study using NS5 also observed results similar to Zhang et al. [] using genome sequence. This study included two additional sequences AMR39830 (KU955589) and AMR39831 (KU955590) deposited in GenBank on 23rd March 2016 hence was not included in the analysis by Zhang et al. []. These two sequences segregated separately in two different clusters (Fig. 1A). However, the amino acid mutation of the Chinese isolate imported from Venezuela at NS5 (AMK79469, KU744693) were not discussed by Zhang et al. []. The substitutions observed were D60E, P62A, H38K, N203I, K38H, and S36A among a total of six substitutions (Table I). Three other sequences analysed were the sequences from Honduras collected in 2016 and Guatemala from fetal brain also in 2016 and a Mexican sequence (ANF04750, Year of collection, 2015). All three sequences show R274C mutation involving cysteine.
Phylogenetic analysis of ZIKV sequences from Brazil was reported by Wang et al. [2016]. The present analysis were built upon the analysis by Wang et al. [2016] using additional sequences. Mutation of E to V was observed in Natal sequence (AMB18850) of NS5 in the present study. However, mutation of V to A in Rio S1 (AMQ48982) of NS5 discussed by the author was not observed in the present analysis. Comparison of substitution at NS5 in the Asian lineages was done with the African lineages. The mutation observed in Asian lineages were N125S, R138H, H198Q, A275I, G336K, R396N, E309K (Table I) which were same as that observed by Wang et al. [2016]. However, the present analysis observed additional mutation R29N, N273S, H383Q, P391S which were not observed by Wang et al. [2016] at NS5 (Table I). Thus, two N to S mutation and two H to Q mutation was found in the analysis. Also, the proline to serine mutation was an interesting observation and it was suggested that the sequence from Philippines, year 2012 (AMD61711) and/or the sequence from Brazil collected in 2015 (AMQ76465) may have reverted the mutation appearing as proline.
The origin of the Asian lineage might be hypothesized to be from African ZIKV based on a partial sequence used in the analysis. Interestingly, the partial sequence of NS5 from an isolate submitted from Australian security CRC, Queensland in 2007, accession no. ABY86749 isolation source unknown (probably isolated from the pacific region), fell within the Ugandan MR766 2006 clade (Fig. 1). The partial sequence was referred to here as the outbreak clade of 2007, the same year the first transmission of Zika virus was reported outside Africa and Asia. The results suggested the outbreak of 2007 from pacific to be the result of introduction of Ugandan strain MR766 into the pacific region probably by travel importation. The partial sequence did not show the mutation at position 29, 125, 138, and 198 (Table I) explaining the African origin.
To conclude the co-circulation of two zika virus genotypes associated with ZIKV was suggested. Based on the results of NS5 sequence analysis the Brazilian genotype and the Latin American genotype of Mexico, Panama, and the Caribbean was proposed. However, it is possible that other sequences depicting co-circulation could have passed unnoticed because of limited sampling and sequencing.
ACKNOWLEDGMENTS
The author wishes to thank Indrani Karunasagar, Director (R&D), Nitte University Centre for Science Education and Research and the Management of Nitte University, Deralakatte, Mangalore, Karnataka, India for the support in establishing the center, providing facilities and continuous encouragement in research, including the present work.




