High prevalence of G12P[8] rotavirus strains in Rio Branco, Acre, Western Amazon, in the post-rotavirus vaccine introduction period
Abstract
The present study aimed to provide a molecular characterization of circulating rotavirus (RVA) strains in Rio Branco, Acre, in the post-rotavirus vaccination period, particularly with regard to the emerging, increasingly prevalent G12P[8] genotype. A total of 488 fecal specimens from diarrheic and non-diarrheic children were obtained between January and December 2012. RVA detection was initially performed using enzyme-linked immunosorbent assay (ELISA) method, followed by reverse-transcription polymerase chain reaction (RT-PCR) using specific primers. RVA was detected in 18.3% (44/241) of the children with acute diarrhea and in 1.2% (3/247) of the non-diarrheic children (P < 0.001), with overall RVA-positivity of 9.6% (47/488). The most common genotype was G2P[4] with 43.2% (19/44) of the diarrheic cases, followed by G12P[8] (27.3%, 12/44), G3P[6] (18.2%, 8/44), G3P[8] (4.5%, 2/44), and G12P[6] (2.3%, 1/44). G12 samples belonged to lineage III and were from children aged 4–52 months. All of these children had acute diarrhea associated with fever (83.3%, 10/12) and vomiting (66.7%, 8/12). Most of the cases occurred in August (58.3%, 7/12), 75% (9/12) of which having received the full vaccination scheme with Rotarix™. For the first time G12 was reported at relative high prevalence in Brazil. Our findings warrant further monitoring studies on the molecular characterization of circulating RVA strains after rotavirus vaccine introduction in Brazil and elsewhere, since the occurrence of either unusual our emerging genotypes may pose a challenge to vaccination strategies. J. Med. Virol. 88:782–789, 2016. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
According to recent estimates group A rotaviruses (RVA) are responsible for more than 197,000 deaths among children under 5 years of age, mostly in developing countries, therefore, representing the most common cause of diarrhea-related hospitalizations and deaths worldwide [Lanata et al., 2013].
Although improvements in sanitation, hygiene practices, and drinking potable water may contribute to the prevention and control of acute diarrheal disease, this clearly do not efficiently prevents the spread of rotaviruses. Consequently, the large-scale use of rotavirus vaccine represents the most effective intervention toward the prevention of severe disease caused by RVA [Tate et al., 2012].
Two oral vaccines containing attenuated RVA strains are already commercially available worldwide. One, called RotaTeq™ (Merck & Co., Whitehouse Station, NJ), is a pentavalent bovine-human vaccine including in its composition G1, G2, G3, G4, and P[8] strains which are the most common human genotypes. The other, Rotarix™ (GlaxoSmithKline Biologicals, Rixensart, Belgium), is a monovalent vaccine derived from the human rotavirus G1P[8] strain [O'Ryan and Linhares, 2009]. Both vaccines are recommended for use by the World Health Organization (WHO) in all countries, especially in those with high diarrhea-related mortality among children younger than 5 years of age [Tate et al., 2012; WHO, 2013]. As a result of the increasing introduction of both rotavirus vaccines in several countries all over the world, a significant reduction in terms of diarrhea-related morbidity and mortality has been observed [Desai et al., 2011; Tate et al., 2012].
Brazil was among the first nations to include Rotarix™ into the routine vaccination schedule. Before the introduction of the RVA vaccine rotavirus accounted for 3.5 million episodes of diarrhea, 650,000 ambulatory care visits, and 92,000 hospitalizations in Brazil. Moreover, rotavirus caused 850 annual deaths among children aged less than 5 years [Sartori et al., 2008]. RVA vaccine was introduced in Brazil in March 2006, just after the more extensive ever reported RVA outbreak in Brazil that has occurred in 2005, in Rio Branco, Acre, Western Amazon, where G9 RVA genotype was responsible for 71% of cases [Linhares and Justino, 2014; Siqueira et al., 2010].
Rotavirus belongs to the Rotavirus genus, Reoviridae family, and Sedoreovirinae sub-family. As based on serological reactivity and genetic variability, currently, addition to the eight groups or species (A–H) recognized, the novel rotavirus species named Rotavirus I has been studied [Matthijnssens et al., 2011; Mihalov-Kovács et al., 2015]. The virion consists of a triple-layered particle surrounding a viral genome with 11 segments of double-strand RNA. Based on the two outer capsid proteins, VP7 and VP4, RVA is classified into G and P genotypes. Currently 27 G and 37 P genotypes are described [Matthijnssens et al., 2011; Trojnar et al., 2013].
Despite the broad genomic and antigenic RVA strain diversity, a limited number of globally prevalent strains have been reported to infect humans over the past three decades. Worldwide, rotavirus strains bearing G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] specificities are knowing to account for about 80–90% of RVA disease burden [Desselberger, 2014]. In addition, G12 RVA strains associated with P[6] and P[8] genotypes are known to be emerging in many settings around the world [Iturriza-Gómara et al., 2011]. Recently G12P[8] strains have emerged at high prevalence rates in several places all over the world [Bányai et al., 2012].
In the past decade, G12 has emerged on a global scale, most often in combination with P[6] or P[8] genotypes and less commonly with P[4] and P[9]. G12 genotype became, therefore, the sixth most prevalent RVA VP7 genotype associated with infections in humans [Rahman et al., 2007; Matthijnssens et al., 2010]. Due to the increasing rise in G12 RVA prevalence rates, it has been postulated that this emerging genotype might even become dominant in the future [Rahman et al., 2007].
The majority of G12 strains have been detected in Asia, more specifically in the southeast part of the continent, which suggests this region as being the likely origin for spreading of G12 strains characterized throughout the world. It has been hypothesized that spread of such strains may have occurred through mobility of humans and animals [Rahman et al., 2007]. An additional hypothesis suggests that emergence of G12 strains might be associated with their potential to escape the immune system unlike it has been recognized common, more prevalent genotypes, including G1–G4 [Samajdar et al., 2006].
G12P[4] combination was first reported in 1987 during an outbreak in Philippines [Taniguchi et al., 1990]. Since then, this genotype has been observed in several other countries. In 2002, G12P[8] was seen to strike the United States [Griffin et al., 2002], Thailand [Pongsuwanna et al., 2002], India [Das et al., 2003], and Japan [Shinozaki et al., 2004]. More recently this genotype was identified in several continents, demonstrating its progressive global propagation over the past decade [Bányai et al., 2012; Stupka et al., 2012; Dóró et al., 2014; Gómez et al., 2014].
There have been some studies in Africa showing that currently available vaccines may offer protection against G12 genotype, even though these vaccines do not contain this specificity in their composition. In the United States, the high efficacy of the RotaTeq vaccine against G12 has been observed (83% [95%CI: 57–93%]) [Payne et al., 2013]. In Africa, efficacy against G12 tended to be lower (51.5% [95%CI: −6.5–77.9%) [Steele et al., 2012].
G12 RVA strains in combination with P[6], P[8], and P[9] have been described in several Brazilian regions [Pietruchinski et al., 2006; Soares et al., 2012, 2014; Gómez et al., 2014]. However, few studies have reported genomic characterization of G12 strains in Brazil [Soares et al., 2012; Gómez et al., 2014].
In this study, we sought to identify potential risk factors related to RVA disease in Rio Branco, Acre, among children aged under 5 years. In addition, we described molecular characterization of G12P[8] strains.
MATERIALS AND METHODS
Patients and Clinical Specimens
From January to December 2012, bimonthly visits were made to Rio Branco, Acre, Northwestern Brazil, to collect fecal specimens from diarrheic and non-diarrheic children younger than 5 years who were either admitted to a local general hospital (collections made up to 48 hr from admission) or sought for treatment at an outpatient unit. For each child, standard questionnaires were filled out with data including living conditions, dietary habits, current disease history, and clinical information. Acute diarrhea was defined as the presence of three or more liquid and semi-liquid stools in a 24-hr period for up to 14 days.
RVA Detection
Stool samples were screened for RVA antigen by ELISA using Ridascreen kit (R-Biopharm®, Darmstadt, Germany) according to the manufacturer's instructions. Viral genome was extracted from 10% fecal suspensions using silica glass powder [Boom et al., 1990]. RNA electrophoretic analysis was performed in vertical polyacrylamide gel electrophoresis (PAGE), as described by Pereira et al. [1983].
RT-PCR and Genotyping
All RVA-positive samples were subjected to reverse transcription-polymerase chain reaction (RT-PCR), using consensus primers Beg9/End9 and 4con3/4con2 to amplify VP7 (1062 bp) and VP4 (876 bp) genes, respectively [Gouvea et al., 1990; Gentsch et al., 1992]. A second round was performed using a set of specific primers for G (G1, G2, G3, G4, G9, and G12) and P-types (P[4], P[6], P[8], and P[9]). All amplified products (amplicons) were visualized in a 1.5% agarose gel.
Nucleotide Sequencing and Phylogenetic Analysis
Sequencing of the PCR amplicons for VP7 and VP4 genes of G12 strains were performed using the same primers as those used in the PCR and carried out with a Big Dye Terminator cycle sequencing kit v 3.1 (Applied Biosystems, Foster City, CA). Electrophoresis was performed in the ABI Prism 3130xl automatic sequencer (Applied Biosystems) and the sequences obtained were aligned and edited using the BioEdit Sequence Alignment Editor program (v. 7.0.5.2). Neighbor-joining method was used to perform the phylogenetic analysis, in which distance was calculated from aligned sequences [Kimura, 1980]. Dendrograms were constructed using the MEGA program v.5.0.1, and bootstrap analysis was performed using 2,000 replicas.
Partial nucleotide sequences of VP7 (931 bp) and VP4 (770 bp) genes from our study were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov), under the following access number: KT025858-KT025868 (VP7 gene) and KT025869-KT025876 (VP4 gene).
Statistical Analysis
The results were processed in a database created with the Statistical Package for Social Sciences (SPSS) v. 20.0 for Windows software. A descriptive statistical analysis was done to obtain relative and absolute frequencies, for both, analyzed and RVA infection prevalence. The association between each of the independent variable with the dependent variable was obtained by means of a chi-square test (χ2). In the multivariable analysis, Poisson regression was performed using the STATA 12.0 program, a method which enabled researchers to evaluate independent variables associated with RVA infection, while controlling for possible confounding factors (adjusted RP). The statistical significance level was set at α = 0.05 (5%) and the confidence interval (IC) was set at 95%.
Ethics
The study was approved by Evandro Chagas Institute's Human Research Ethics Committee, protocol number 526.784, in accordance with National Health Council's Resolution 466/2012. The authors ensure that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
RESULTS
In January, March, June, August, and November 2012, a total of 488 fecal samples were collected from children aged between 0 and 5 years old, of whom 241 presented with acute diarrhea and 247 were non-diarrheic patients. Overall RVA-positivity was 9.6% (47/488). RVA-positivity rates of 18.3% (44/241) and 1.2% (3/247) were recorded among diarrheic and non-diarrheic children, respectively (P < 0.001).
RVA-positive samples were subjected to PAGE and typical RVA electrophoretic pattern was displayed in 63.8% (30/47) of positive samples. Short and long electropherotypes were seen in 73.3% (22/30) and 26.7% (8/30) of specimens, respectively. RVA strains bearing G12P[8]-specificity displayed long RNA profile in 7 (58.3%) of the 12 strains that were electropherotyped strains (data not shown).
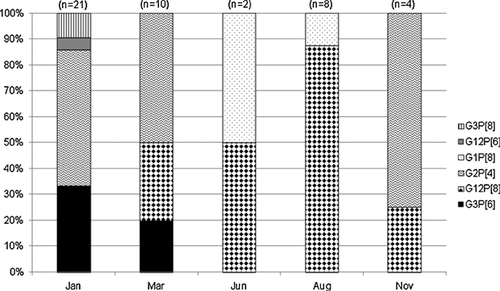
G and P type-specificities could be assigned to 95.7% (45/47) of samples. The usual most commonly detected genotypes from diarrheic children were G2P[4] (42.3%, 19/45), G12P[8] (26.7%, 12/45), and G3P[8] (4.4%, 2/45). In addition, genotypes generally characterized as unusual were found in 20.4% (9/45) of cases, including types G3P[6] (17.8%, 8/45) and G12P[6] (2.2%, 1/45) (Fig. 1). The genotypes identified in samples of non-diarrheic children were G1P[8] (4.4%, 2/45) and G3P[6] (2.2%, 1/45).
Eleven VP7 and eight VP4 sequences of G12P[8] were obtained. Phylogenetic analysis of VP7 gene showed that G12 samples from Acre had high nucleotide (99–100%) and amino acid (98–100%) similarities among themselves. This analysis also showed that our G12 strains grouped into lineage III that included G12 Brazilian strains (Fig. 2A). With regards to VP4 gene, partial analysis demonstrated that G12P[8] strains were 100% similar to each other, also denoting a high homology with a strain isolated in Paraguay in 2008 (1670SR); altogether these strains clustered into lineage III, as shown in Figure 2B. High nucleotide and amino acid sequence similarities (97% and 92%, respectively) were observed between P[8] RVA strains when compared with a cluster of G9P[8] strains isolated during an outbreak in Rio Branco, Acre, Brazil in 2005. In contrast, relatively lower amino acid similarities were observed when comparing our P[8] strains with RotarixTM and RotateqTM vaccines: 75% and 73%, respectively (data not shown).

Among rotavirus-positive patients 80.9% (38/47), 59.6% (28/47), and 93.6% (44/47) had developed fever, vomiting, and diarrhea, respectively. Diarrhea only and diarrhea associated with fever and vomiting were recorded in 5 (11.6%) and 24 (55%) out of 44 patients, respectively. Diarrhea, vomiting, and fever were significantly more common among rotavirus-positive patients, as compared with rotavirus-negative patients (Table I).
| Rotavirus | |||
|---|---|---|---|
| Symptoms | Positive | Negative | P-Value |
| Diarrhea | 93.6% (44/47) | 44.7% (197/441) | <0.001 |
| Vomiting | 59.6% (28/47) | 15.2% (67/441) | <0.001 |
| Fever | 80.9% (38/47) | 34.2% (151/441) | <0.001 |
General characteristics of rotavirus-positive cases are summarized in Table II. The median age of children was 21 months and most of the subjects (51.1%, 24/47) gathered within the 6–24 month age group. As to the gender, 61.7% of children were male. Exclusive breast-feeding during the first 6 months of life was reported for 14.9% (7/47) of these children. No statistically significant differences (P > 0.2) were seen when comparing age groups, genders, and feeding types.
| Characteristics | Cases, n (%) | P-Value |
|---|---|---|
| Age groups | ||
| <6 months | 8 (17) | |
| 6–24 months | 24 (51.1) | 0.715 |
| >24 months | 15 (31.9) | |
| Gender | ||
| Male | 29 (61.7) | 0.259 |
| Female | 18 (38.3) | |
| Exclusive breastfeeding | ||
| Yes | 7 (14.9) | |
| No | 28 (59.6) | 0.337 |
| Not reported | 12 (25.5) |
A higher distribution (58.3%, 7/12) of G12P[8] strains was observed in August 2012 and were detected in children aged between 4 and 52 months. All of these children presented acute diarrhea associated with fever (83.3%, 10/12) and vomiting (66.7%, 8/12) (data not shown). Among G12P[8] rotavirus-positive children 58.3% (7/12) of them had fever, vomit, and diarrhea, demonstrating the severity of the infection.
Multivariate analyses were assessed to show a possible relationship between social, environmental, and household conditions and the risk of becoming infected by rotavirus. The following potential risk factors were less likely to be associated with rotavirus infection: water consumption from the municipal water supply (Prevalence ratio (Pr) 1.85 CI 95% 1.02–3.37; P 0.042) and children with either no rotavirus vaccination or incomplete vaccination schema (Pr 2.29 CI 95% 1.32–3.95; P 0,003), as shown in Table III. Lack of proper waste disposal correlated with a higher likelihood of becoming infected by rotavirus (Pr 2.14 CI 95% 1.08–4.24; P 0.028).
| Variable | RPbrute (CI95%) | P-Value | RPadjusted (CI95%) | P-Value |
|---|---|---|---|---|
| 2° Blocka | ||||
| Access to water supply | ||||
| Municipal water supply | 1.55 (0.88–2.73) | 0.132 | 1.85 (1.02–3.37) | 0.042 |
| Well water/river/igarape | 1 | 1 | ||
| Waste disposal | ||||
| Outside toilet /river/igarape | 1.79 (0.94–3.44) | 0.078 | 2.14 (1.08–4.24) | 0.028 |
| Sewerage/septic tank | 1 | 1 | ||
| 3° Blockb | ||||
| Rotavirus vaccination status | ||||
| None or one dose | 2.05 (1.19–3.52) | 0.009 | 2.29 (1.32–3.95) | 0.003 |
| Two doses | 1 | 1 |
- RPbrute, variables not adjusted - bivariate analysis. CI 95%, 95% confidence interval. P, descriptive level of association of χ2 test. RPadjusted, variables adjusted between each other - multiple analysis.
- a 2° Block, variables adjusted between each other, because Block 1 had not adjusted to the variable model.
- b 3° Block, variables adjusted between each other and by Block 3.
DISCUSSION
The present study shows a RVA prevalence of 18.3% (44/241) among diarrheic children in Rio Branco, Acre, Western Amazon in 2012. This positivity rate appears lower than those (18.8–37%) from other studies throughout the world [Espejo et al., 2014; Tort et al., 2015]. In Brazil, rotavirus prevalence has been shown to vary over time and to decrease after rotavirus vaccine introduction. Between 2002 and 2011, for instance, RVA prevalence rates were found to decline from 11.2% to 5.07% if pre- and post-vaccination periods are compared [Assis et al., 2013]. Studies in the country's northern region have shown RVA positivity rates in the range of 20–71% [Justino et al., 2011; Soares et al., 2014].
In the present study, G2P[4] accounted for 40.4% of diarrheic children, possibly reflecting findings from several other studies across Brazil and elsewhere which showed a predominance of this genotype in several settings during the first years after introduction of rotavirus vaccination [Justino et al., 2011; Kirkwood et al., 2011]. It has been postulated that such a predominance of G2 strains might be due to a vaccine-induced selective pressure, even though a fluctuation over time cannot be ruled out. In favor of this latter hypothesis, there have been reports on the dominance of G2P[4] genotype even in countries where universal rotavirus vaccination was not yet introduced [Linhares et al., 2011; Oliveira et al., 2012].
The globally emerging G12P[8] genotypes were second to G2P[4], accounting for 23.4% of isolates in Rio Branco in 2012. G12 strains are reported to have emerged globally during the past decade, and more recently these VP7 genotypes were reported in the Northern region of Brazil in the early post-rotavirus vaccination period [Soares et al., 2012]. This represents the first study in Brazil that reports a high prevalence of G12 (27.6%, 13/47) associated with P[8] and P[6], when compared to earlier studies conducted all over Brazil [Gómez et al., 2014; Soares et al., 2014]. The global spread of G12 strains might potentially pose a challenge to rotavirus vaccination strategies, since current available vaccines do not contain this specific type into their compositions. Nonetheless, a few vaccine trials conducted in Africa have shown that heterotypic protection against this specific strains may be achieved by vaccination [O'Ryan and Linhares, 2009; Yen et al., 2011].
G12P[8] strains have been reported in Argentina (at 24.8% prevalence rate, between 2008 and 2009), the United States (4%, between 2005 and 2008), and Peru (9.4%, between 2010 and 2012), highlighting the wide spread of this genotype on a global scale [Hull et al., 2011; Stupka et al., 2012; Espejo et al., 2014].
G12P[8] strains have been detected at variable rates (3.2% through 57%) in India, Ethiopia, Nepal, and the northern region of Spain, particularly among diarrheic children in the 6–24 month age group. A predominance of G12P[8] was also observed in this age group in our study, although this may only reflect the high prevalence rate of rotavirus infection in general and not of specific genotypes [Samajdar et al., 2006; Cilla et al., 2013; Kang et al., 2013; Abebe et al., 2014].
Analysis of VP7 genes showed that G12P[8] samples grouped into lineage III denoting a high nucleotide sequence similarity (>98%) among themselves. Noticeably, recent studies have demonstrated that current circulating strains belong to lineage III [Rahman et al., 2007; Matthijnssens et al., 2010; Gómez et al., 2014; Soares et al., 2014].
In our study, the unusual G3P[6] genotype was also found to account for a significant proportion (∼20%) of the isolates, yielding a prevalence rate similar to that reported by Soares et al. [2014], who reported this genotype as being associated with 15% of RVA infections in the Western Amazon region between 2011 and 2012, particularly in Amazonas and Acre states. Recently, G3P[6] has been reported in Argentina at a positivity rate of 6.3%, an apparent much higher prevalence than that for Latin America as a whole, <1% [Linhares et al., 2011].
Our study showed a higher RVA prevalence rate (51%) among children aged 6–24 months, a finding consistent with most of the previously published studies. Studies conducted in the post-rotavirus vaccine era have suggested a shift of higher prevalence rates to older age groups children reaching the ages of 2–5, possibly due to vaccination occurring at an age younger than 6 months [Linhares et al., 2011; Linhares and Justino, 2014].
The wide dissemination of RVA denotes effective inter-human transmission facilitated by its recognized high stability in the environment. The epidemic of acute gastroenteritis caused by RVA G9P[8] in Rio Branco, Acre, Brazil, in 2005, a pre-vaccination time-point period, was significantly associated with the municipality water supply for domestic use [Siqueira et al., 2010]. Although this might have happened by chance, the present study showed that 63.8% of cases had access to the municipality water supply system (P 0.042), whereas the remainder of the cases of (21.3%) lacked proper sewage facilities (P 0.028). These variables were positively associated with the occurrence of RVA in Rio Branco, Acre.
In conclusion, our data improved our knowledge on the diversity of circulating RVA strains in the Amazon region and clearly showed the current prominent role of G12P[8] as a cause of childhood diarrhea in Rio Branco, Acre. It is worth highlighting the importance of a continuous surveillance for circulating RVA strains in the Amazon region of Brazil and elsewhere, since this may be beneficial in determining the overall impact of rotavirus vaccines.
ACKNOWLEDGMENTS
The authors wish to thank all Urgent Care Units and Family Health Posts that took part in this study, particularly to physicians Flávia Fernandes and Guilherme Pulici, as well as all the children and those responsible for their care. Thanks also to the Evandro Chagas Institute for financial and technico-scientific support for the execution of this work. Finally, thanks are due to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for awarding masters’ scholarships.




