Real-time polymerase chain reaction for diagnosing infectious mononucleosis in pediatric patients: A systematic review and meta-analysis
Abstract
In this meta-analysis, we evaluated the diagnostic role of Epstein-Barr virus deoxyribonucleic acid detection and quantitation in the serum of pediatric and young adult patients with infectious mononucleosis. The primary outcome of this meta-analysis was the sensitivity and specificity of Epstein-Barr virus (EBV) deoxyribonucleic acid (DNA) detection and quantitation using polymerase chain reaction (PCR). A systematic review and meta-analysis was performed by searching for articles that were published through September 24, 2014 in the following databases: Medline, Cochrane, EMBASE, and Google Scholar. The following keywords were used for the search: “Epstein-Barr virus,” “infectious mononucleosis,” “children/young adults/infant/pediatric,” and “polymerase chain reaction or PCR.” Three were included in this analysis. We found that for detection by PCR, the pooled sensitivity for detecting EBV DNA was 77% (95%CI, 66–86%) and the pooled specificity for was 98% (95%CI, 93–100%). Our findings indicate that this PCR-based assay has high specificity and good sensitivity for detecting of EBV DNA, indicating it may useful for identifying patients with infectious mononucleosis. This assay may also be helpful to identify young athletic patients or highly physically active pediatric patients who are at risk for a splenic rupture due to acute infectious mononucleosis. J. Med. Virol. 88:871–876, 2016. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Patients with infectious mononucleosis often present with pharyngitis, cervical lymph node enlargement, fever, and fatigue, and 50% to 90% of adults are seropositive for the Epstein-Barr virus (EBV) [Cheng et al., 2007; Sakamoto et al., 2012; Balfour et al., 2015]. Some cases with infectious mononucleosis are secondary to human cytomegalovirus [Raja and de Quesada, 2015]. Infectious mononucleosis is more commonly observed in children and young adolescents due to most adults have already developed immunity to EBV because of prior exposure.
To diagnose infectious mononucleosis secondary to EBV, assays are used that test for antibodies to EBV capsid or EBV nuclear antigens. These assays have high sensitive and specificity, but are expensive [Bell et al., 2006]. Although the heterophile antibody have similar specificity and cost less, the tests are less sensitive in children and adults during the first few days of the illness [Cheng et al., 2007]. In fact, heterophilic antibody tests have a 25% false-negative rate during the first week of acute infectious mononucleosis [Womack and Jimenez, 2015]. During an acute infection or the lytic phase of the infection, viral DNA can also be detected in serum [Bauer et al., 2005]. A polymerase chain reaction (PCR)-based assay that detects EBV DNA has been shown to more sensitive and have higher specificity than that of heterophile antibody tests in children [Bell et al., 2006]. A study by Bauer et al. found detection of EBV DNA by PCD had a sensitivity of 94.9% and a specificity of 97.4% for primary EBV infection [Bauer et al., 2005].
The purpose of this meta-analysis and systematic review was to evaluate the diagnostic role of using PCR to detect and quantitate the level of EBV DNA in the serum of pediatric patients with infectious mononucleosis. The primary outcome of this meta-analysis was the sensitivity and specificity of EBV DNA detection and quantitation using PCR.
MATERIALS AND METHODS
Search Strategy
A systematic review and meta-analysis was conducted by searching for articles published through September 24, 2014 in the following databases: Medline, Cochrane, EMBASE, and Google Scholar. We used the following keywords for this search: “Epstein-Barr virus,” “infectious mononucleosis,” “children/young adults/infant/pediatric,” and “polymerase chain reaction or PCR.” Reference lists of relevant studies were also hand-searched.
Selection Criteria
The inclusion criteria were: (i) subjects of the study had to be children, adolescents and/or younger adults aged <26 years old; (ii) the studies had to use quantitative real-time PCR (RT-PCR) for detection; (iii) the studies had to either report sensitivity and specificity, or provide sufficient data to calculate sensitivity/specificity; and (iv) the studies had to be published after 1992 when RT-PCR became available. Letters, comments, editorials, case reports, proceedings, and personal communications were excluded. We also excluded studies if the outcomes (sensitivity and specificity) were not described or could not be derived from reported data.
Study Selection and Data Extraction
Two independent reviewers identified the studies using the above mentioned search strategy. If any uncertainty regarding eligibility existed, a third reviewer was consulted. The following information and data were extracted from studies that met the inclusion criteria: first author's name, publication year, study design, number of participants in each treatment group, participants’ age and gender, type of RT-PCR used, commercial kits used to extract DNA, DNA source, cut-off values, and sensitivity and specificity (or data available to calculate sensitivity and specificity, i.e., true positive, true negative, false positive, and false negative).
Outcome Measures
For this meta-analysis, we used the following outcomes for analysis and comparison: sensitivities, specificities, and odds ratios (OR) which were either reported or calculated from available data (such as the number of samples in each true positive, true negative, false positive, and false negative category).
Statistical Analysis
For the statistical analysis, we calculated pooled measures of diagnostic performance, such as sensitivity, specificity, and diagnostic odds ratios (DORs), with their corresponding 95% confidence intervals (95%CIs). We also calculated the area under the receiver-operating characteristic (ROC) curve (AUC). The DORs combined sensitivity and specificity into one measure for diagnostic performance. A DOR of one implied that the test had no ability to discriminate between the presence or absence of EBV DNA. For this study, the higher the DOR, the better the diagnostic accuracy of EBV DNA the better the test was for detecting infectious mononucleosis in pediatric patients [Reitsma et al., 2009]. To test for homogeneity, a χ2 test was conducted, and an inconsistency index (I2) statistic was determined. If the I2 statistic was >50% or >75%, the studies were considered to be heterogeneous or highly heterogeneous, respectively. If I2 was <25%, the studies were considered to be homogeneous. If the I2 statistic was >50%, a random-effects model (DerSimonian-Laird approach) was calculated. Otherwise, fixed-effects model (Mantel-Haenszel approach) was used. All statistical assessments were two-sided, and a P value of <0.05 was considered significant. All analyses were performed using Meta-Disc (version 1.4) [Zamora et al., 2006].
RESULTS
Literature Search
During a database search, 195 articles were found (a study selection flowchart is shown in Fig. 1A). After removing the duplicates, 165 studies were assessed, and a total of 36 articles were found eligible. Of these articles, 17 were excluded because they either met one of the exclusion criteria or did not meet all of the inclusion criteria. As for the remaining 19 articles, 15 were excluded after a full-text review because they did not include the outcomes data assessed in this analysis. For this meta-analysis, a total of four studies were included in this analysis.
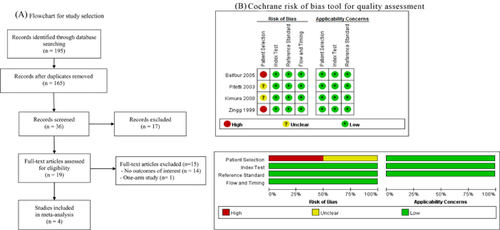
Quality Assessment
The quality of the included studies was evaluated using the assessment of diagnostic studies (QUADAS-2) tool [Whiting et al., 2011]. The quality was judged by two reviewers independently, with a third reviewer consulted for any discrepancy. The results of quality assessment of the included studies are shown in Figure 1B.
Study Characteristics
A total of four studies were included in the analysis [Zingg et al., 1999; Kimura et al., 2000; Pitetti et al., 2003; Balfour et al., 2005]. The characteristics of the studies are summarized in Table I. The number of patients in the included studies ranged from 18 to 79 (total = 159). The patients’ mean age was fairly consistent among the studies at approximately 9 years old, except for Balfour et al. [2005] which examined college students. In most studies, excluding Balfour et al., there were slightly more males than females.
| 1st AU | Number of pediatric or young adult patients | Agea (y) | Male (%) | Types of PCR detection used | Kits used to extract DNA | Extracted DNA source |
|---|---|---|---|---|---|---|
| Balfour et al. [2005] | 25 | 20.6 (18–26) | 28% | TaqMan PCR assay; real-time quantitative | QIAamp DNA Blood Mini Kit (Qiagen) | Whole blood |
| Pitetti et al. [2003] | 79 | 9y 3 mo | 54% | TaqMan PCR assay; real-time quantitative | QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA) | Serum |
| Kimura et al. [2000] | 37 | 4 (1–8) for IM group, 6 (8 mo-16) for control group | NR | TaqMan PCR assay; real-time quantitative | QIAamp Blood Kit (Qiagen, Chatsworth, CA) | Serum |
| Zingg et al. [1999] | 18 | 8.6 | 56% | Real-time quantitative PCR | QIAamp blood kit (Qiagen, Basel, Switzerland) | Serum |
- AU, author; y, year; mo, month; NR, not reported; PCR, polymerase chain reaction; TP, true positive; TF, true negative; FP, false positive; FN, false negative.
- a Numbers within parenthesis were range.
Sensitivity, Specificity, Diagnostic Odd Ratios, and Area Under the Receiver-Operating Characteristic Curve (AUC) for All Patients
The sensitivity and specificity for EBV DNA in the four studies are listed in Table II, and ranged from 75.0% to 81.5% and 82.1% to 100%, respectively [Zingg et al., 1999; Kimura et al., 2000; Pitetti et al., 2003; Balfour et al., 2005]. There was no significant heterogeneity regarding sensitivity among the studies (Q = 0.61; I2 = 0%; P = 0.894). Therefore, a fixed-effect model was used for this meta-analysis. The pooled sensitivity for EBV DNA was 80% (95%CI, 73–85%) (Fig. 2A).
| 1st AU | Number of pediatric or young adult patients | Cutoff value (copies/ml) | Sensitivity (%) | Specificity (%) | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|
| Balfour et al. [2005] | 25 | 10 | 81.5 | 82.1 | 75 | 5 | 17 | 23 |
| Pitetti et al. [2003] | 79 | 10 | 75 | 98 | 21 | 1 | 7 | 50 |
| Kimura et al. [2000] | 37 | 2 | 78.8 | 96.4 | 26 | 1 | 7 | 27 |
| Zingg et al. [1999] | 18 | 10 | 77.8 | 100.0 | 14 | 0 | 4 | 29 |
- AU, author; NR, not reported; PCR, polymerase chain reaction; TP, true positive; TF, true negative; FP, false positive; FN, false negative.
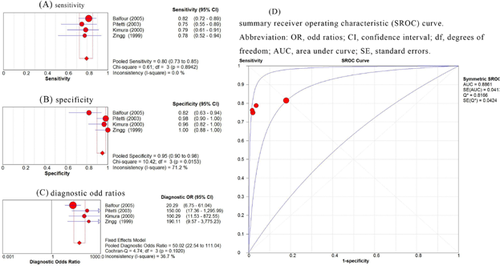
For sensitivity, the homogeneity tests demonstrated that the Q statistic was 10.42 and that the I2 statistic was 71.2% (P = 0.015). As these results demonstrated that significant heterogeneity existed among the studies, a random-effects model was used for the pooled specificity. The pooled specificity for EBV DNA was 95% (95%CI, 90–98%) (Fig. 2B).
For DOR, significant homogeneity existed among the studies with a Q statistic of 4.74 (P = 0.192) and an I2 statistic of 36.7%, as assessed by inspecting a Forest plot (Fig. 2C). The pooled DOR across the studies was 50.02 (95%CI, 22.54–111.04). The AUC for the ROC curves was 0.8861 (with a standard error of 0.041; Fig. 2D). The high DOR value and the closeness of the ROC AUC to one indicates that the PCR test has high accuracy for diagnosing infectious monucleosis in this patient group accurately.
Sensitivity, Specificity, Diagnostic Odd Ratios, and Area Under the Receiver-Operating Characteristic Curve (AUC) for Children Only
We also performed the analysis in which the study that included college age students was omitted [Balfour et al., 2005] from the analysis, leaving only those studies with pediatric patients. There was no significant heterogeneity regarding sensitivity among the studies (Q = 0.13; I2 = 0%; P = 0.939). Therefore, a fixed-effects model was used for this meta-analysis. The pooled sensitivity for EBV DNA was 77% (95%CI, 66–86%) (Fig. 3A).
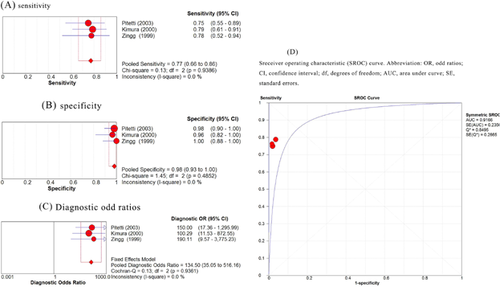
The homogeneity tests for specificity demonstrated that the Q statistic was 1.48 and that the I2 statistic was 0% (P = 0.485), indicating significant homogeneity existed among the studies. Hence, a fixed-effects model was used for the pooled specificity. Figure 3B shows that the pooled specificity for EBV DNA was 98% (95%CI, 93–100%).
For all studies, the pooled DOR was 134.50 (95%CI, 35.05–516.16), with a Q statistic of 0.13 for heterogeneity (P = 0.936) and an I2 statistic of 0%. Significant homogeneity existed among the studies, as assessed by inspecting a Forest plot (Fig. 3C). The area under the summary ROC curves, or AUC was 0.9166 (with a standard error of 0.235; Fig. 3D). Based on the DOR value and the summary ROC curves, EBV DNA could be used to assess infectious monucleosis in pediatric patients even more accurately than in younger adult patients.
Publication Bias
An analysis for publication bias was not performed because more than six studies are required to detect funnel plot asymmetry for the primary outcome [Sterne et al., 2011].
DISCUSSION
Infectious mononucleosis occurs most commonly in patients aged 15–24 years [Vouloumanou et al., 2012]. Moreover, infectious mononucleosis secondary to EBV can sometimes be recognized by a triad of symptoms including fever, sore throat, and lymphadenopathy [Vine et al., 2012]. As for diagnostic testing, EBV DNA has been found in blood lymphocytes of 54–94% of healthy seropositive persons [Chan et al., 2001]. To diagnose infectious mononucleosis secondary to EBV, testing for antibodies to EBV capsid or Epstein-Barr nuclear antigens are the most accurate methods but these methods are expensive [Bell et al., 2006]. A prior study by Yamamoto et al. demonstrated that 100% of patients with acute infectious mononucleosis had EBV DNA in their serum, whereas only 44% of patients had EBV DNA in their serum during convalescence [Yamamoto et al., 1995]. Although heterophile antibody tests have a similar specificity as the tests that use antibodies to detect the presence of the EBV capsid or Epstein-Barr nuclear antigens, and have less cost, they are less sensitive in children and adults during the first few days of the illness [Bell et al., 2006]. To evaluate the diagnostic role of PCR-based assays for detecting EBV DNA in the serum of pediatric and young adult patients with infectious mononucleosis, we conducted a meta-analysis and systematic review. The primary outcome of this meta-analysis was the sensitivity and specificity of EBV DNA detection and quantitation using PCR.
For the four assessed studies in this meta-analysis, the range in sensitivity and specificity for detecting EBV DNA was 75.0% to 81.5% and 82.1% to 100%, respectively [Kimura et al., 1999; Zingg et al., 1999; Pitetti et al., 2003; Balfour et al., 2005]. We found that the pooled sensitivity for PCR to detect EBV DNA was 77% (95%CI, 66–86%), and the pooled specificity was 98% (95%CI, 93–100%). We also found that the accuracy for diagnosing mononucleosis in this patient population was high as evaluated by DOR and the ROC AUC.
Our findings are consistent with prior studies A by Fafi–Kremer et al. demonstrated a specificity of 100% and a sensitivity of 58% for PCR to detect EBV DNA in the plasma of 100 EBV-seropositive persons who had acute infectious mononucleosis, EBV-related diseases secondary to human immunodeficiency virus, or healthy EBV-seropositive persons using PCR detection [Fafi-Kremer et al., 2004]. Yamamoto et al. demonstrated that the detection of EBV DNA by PCR had a specificity of 100% during acute infectious mononucleosis and a sensitivity of 44% during convalescence with PCR in 20 patients with infectious mononucleosis [Yamamoto et al., 1995]. In a study reported by Bauer et al. with 98 patients who were immunocompetent and who had serologically confirmed primary EBV infection, detection PCR tests for EBV DNA had a sensitivity of 94.9% and a specificity of 97.4% for a primary EBV infection [Bauer et al., 2005].
As for a quality assessment of the three studies included in this meta-analysis, the pooled DOR was 102.20 (95%CI, 22.47–464.86). Further, the Q statistic was 1.01 for heterogeneity (P = 0.602). The I2 statistic was 0%. By inspecting a Forest plot, significant homogeneity existed among the studies.
This meta-analysis is one of the few analyses and perhaps the only meta-analysis to examine PCR detection for EBV DNA in the serum of patients with acute infectious mononucleosis. Given the pooled sensitivity for EBV DNA of 77% (95%CI, 66–86%) and pooled specificity for EBV DNA of 98% (95%CI, 93–100%), PCR may serve as a supplementary confirmation method to heterophile antibody tests that have had a reported 25% false-negative rate during the first week of illness, as reported in a prior study [Womack and Jimenez, 2015]. Furthermore, PCR enhances diagnostic accuracy and can be important for patients who participate in contact sports or heavy physical activity and are at risk of splenic rupture.
This meta-analysis has some limitations. One limitation involved the limited number of available studies to include in this meta-analysis. Another limitation was that the majority of studies were published a long time ago, with two published in 1999 and one published in 2003. Further, the clinical utility of this meta-analysis was unclear because PCR is costly and more time consuming to perform, especially for a mostly self-limiting disease such as infectious mononucleosis. Lastly, publication selection bias could not be assessed because of the small number of studies included in this analysis; to assess publication selection bias, more than six studies are necessary [Sterne et al., 2011]. Therefore, additional study and analysis of EBV DNA detection for infectious mononucleosis is necessitated to alleviate these limitations.
This meta-analysis demonstrated that EBV DNA may be helpful in the diagnosis of infectious mononucleosis secondary to EBV using PCR detection methods. Based on the results of our analysis, PCR-based tests had a lower sensitivity but a higher specificity than then the heterophile antibody test, and hence, may be an alternative detection method and/or a confirmatory test for diagnosing mononucleosis. Although infectious mononucleosis secondary to EBV is mostly a self-limiting illness, PCR detection methods for EBV DNA could help in the diagnosis of young athletic or highly physically active pediatric patients who have a risk of splenic rupture with infectious mononucleosis. Moreover, PCR detection of EBV DNA could be useful for pediatric patients who had an organ transplant because of their decreased immunity and higher risk of EBV infection.




