Molecular epidemiology of Crimean–Congo hemorrhagic fever in Bulgaria—An update
Abstract
Crimean–Congo hemorrhagic fever (CCHF) is endemic in Bulgaria. During 2013–2014, 11 confirmed CCHF cases have been reported in the country (seven in 2013 and four in 2014). The present study provides the CCHF molecular epidemiology in Bulgaria based on all currently available S, M, and L RNA segment nucleotide sequences spanning the years 1978–2014. A relatively low genetic difference (0–6%, the maximum seen in the M RNA segment) was seen among the CCHFV sequences suggesting that a slow evolving CCHFV strain belonging to “Europe 1” clade is present in Bulgaria. Although the virus emerged in new foci during the recent years, it is more active in the established endemic foci which seem to offer the most suitable ecosystem and environment. Understanding the CCHF epidemiology and virus evolution is the basis for public health programs and vaccine design. J. Med. Virol. 88:769–773, 2016. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Crimean–Congo hemorrhagic fever (CCHF) is a potentially severe disease characterized by fever and hemorrhagic manifestations. It is caused by CCHF virus (CCHFV) (genus Nairovirus, family Bunyaviridae), a negative-stranded RNA virus with a 3-segmented genome: small (S), medium (M), and large (L) segments, encoding the nucleocapsid protein, the glycoprotein precursor (giving rise to Gn and Gc glycoproteins), and the RNA-dependent RNA polymerase, respectively [Schmaljohn and Nichol, 2007]. Based on the S RNA segment sequences, CCHFV strains can be classified into at least six distinct genetic lineages, which present an approximate geographic distribution [Hewson et al., 2004; Deyde et al., 2006]. Humans are infected through bites of infected ticks (mainly Hyalomma spp.) or by direct contact with blood or tissues from viremic patients or animals [Papa, 2010]. CCHF endemic foci are present in Asia, Africa, and Europe, with the disease distribution coinciding with that of the Hyalomma spp. ticks. Regarding the Balkan peninsula, outbreaks or sporadic cases have been reported in Albania, Bulgaria, and Kosovo, whereas one fatal case was reported in 2008 in Greece [Papa et al., 2002a, 2002b, 2004, 2010; Duh et al., 2007]. Two CCHFV lineages have been reported in Balkans: “Europe 1” (or V) and “Europe 2” (or VI). “Europe 1” lineage contains pathogenic strains from Balkans, Turkey, and Russia, whereas “Europe 2” lineage contains the Greek strain AP92 which is considered of having low pathogenicity [Papa et al., 2015]. Additional AP92-like strains have been detected recently in Turkey, Kosovo, and Greece [Midilli et al., 2009; Papa et al., 2014; Sherifi et al., 2014].
Located on the Balkan Peninsula in southeastern Europe, Bulgaria is endemic for CCHF. The disease was first described in the country in 1952 [Nekliudov, 1952]. A formalin-inactivated mouse brain-derived vaccine is applied in groups of professionally exposed people (e.g., military personnel, veterinarians) in the endemic regions [Papa et al., 2011; Mousavi-Jazi et al., 2012]. The number of CCHF cases decreased in the last years (less than 10 cases annually), which can be attributed to the vaccination, but also to additional important factors, like better information of the people about the risk and the protection measures, and the lower number of persons engaged in rural activities resulting in decreased exposure to ticks.
In total, 70 CCHF cases have been reported during the last decade (2005–2014); 11 (15.7%) were fatal. Most of the cases originated from southern Bulgaria, mainly from southeastern regions of the country, not far from the borders with Turkey and Greece. Since 2009, new foci have been identified (southeastern, northeastern, and south-central districts of Bulgaria) [Christova et al., 2009, 2013].
The first phylogenetic study on Bulgarian CCHFV sequences from strains isolated during 2002–2003 showed that they constitute a distinct clade in the “Europe 1” lineage [Papa et al., 2004]. Additional sequences have been obtained in the following years [Papa et al., 2004, 2011; Christova et al., 2009; Lumley et al., 2014] and unpublished data. The aim of the present study was the genetic detection and isolation of CCHFV in samples from human cases which occurred in Bulgaria in 2013–2014 and the molecular epidemiology of CCHF in Bulgaria based on all currently available S, M, and L RNA segment nucleotide sequences.
MATERIALS AND METHODS
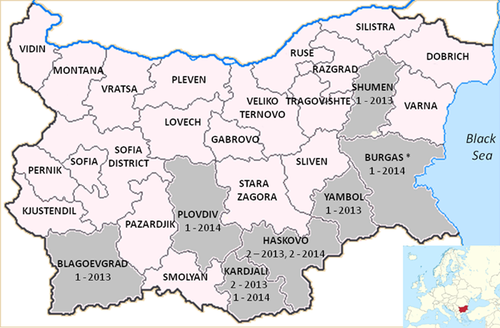
During 2013–2014, 11 confirmed CCHF cases have been reported in Bulgaria, seven in 2013 and four in 2014 (Table I). The median age of the patients was 56 years (range 10–75 years). Nine were male. Three cases (27.3%) were fatal. All, except one, had documented history of recent tick bite. Four cases occurred in Haskovo, three in Kardzhali, and one each in Blagoevgrad, Shumen, Plovdiv, and Yambol (Fig. 1). The laboratory diagnosis was performed at the National Centre for Parasitic Diseases in Sofia, Bulgaria. A commercially available ELISA kit (Vector Best, Russia) was used to detect CCHFV IgM and IgG antibodies, while a real-time RT-PCR [Garrison et al., 2007] and a nested RT-PCR [Schwarz et al., 1996] were used to detect CCHFV RNA. Blood samples which were PCR-positive in Bulgaria (five cases of 2013), together with cDNA from two PCR-positive cases of 2014, were sent to Aristotle University of Thessaloniki for further analysis.
| ELISA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Sex | Age | District | Month | Day of illness | Tick bite | IgM | IgG | Viral load copies/µl | Hemorrhage/outcome |
| V13/13a | M | 39 | Kardzhali | Mar | 9 | No | neg | neg | 1.3 × 103 | Yes/fatal |
| V32/13 | M | 50 | Yambol | May | 7 | Yes | pos | neg | 3.8 × 104 | Yes |
| V36/13 | F | 63 | Blagoevgrad | May | 10 | Yes | pos | pos | 2.5 × 105 | Yes |
| V41/13 | M | 17 | Shumen | Jun | 12 | Yes | grey zone | pos | 1.8 × 103 | Yes |
| V46/13 | M | 58 | Haskovo | Jun | 4 | Yes | neg | neg | 4.5 × 107 | Yes/fatal |
| V64/13 | M | 59 | Haskovo | Jul | 10 | Yes | pos | pos | - | Yes |
| V120/13 | F | 59 | Kardzhali | Nov | 39 | Yes | neg | pos | - | Yes |
| V19/14 | M | 75 | Plovdiv | Apr | 7 | Yes | grey zone | neg | 3.8 × 105 | Yes/fatal |
| V85/14 | M | 56 | Haskovo | Jul | 9 | Yes | neg | neg | 1.62 × 106 | Yes |
| V16/14 | M | 10 | Kardzhali | Apr | 18 | Yes | pos | pos | - | Yes |
| V21/14 | M | 42 | Haskovo | May | 15 | Yes | pos | pos | - | Yes |
- The viral load of the seven cases included in the present study is shown.
- a Co-infection with Plasmodium falciparum.
Viral RNA was extracted from the blood samples using the QIAamp Viral RNA Mini kit (Qiagen GmbH, Hilden, Germany). A quantitative real time RT-PCR and a nested RT-PCR were applied [Schwarz et al., 1996; Papa et al., 2007]. Amplicons were bi-directional sequenced using the BigDye Terminator Sequencing Kit on a 3130 Genetic Analyzer (Life Technologies, Carlsbad, CA). Phylogenetic analysis was performed using the MEGA6 software [Tamura et al., 2013]. Virus isolation was attempted from the blood sample with the highest viral load. A 10% suspension in phosphate-buffered saline was inoculated into a T25 flask containing confluent Vero E6 cells. The supernatant of the culture was checked by the quantitative real time RT-PCR [Papa et al., 2007].
RESULTS
The CCHF viral load was measured in blood samples from seven CCHFV cases observed in Bulgaria during 2013–2014. It ranged from 1.3 × 103 to 4.5 × 107 copies/ml (Table I). A CCHFV strain (Haskovo strain) was isolated from the sample with the highest viral load (V46/13). The primary isolate was passaged three times. The RT-nested PCR was positive in all cases except one (V41/13). Apart the six sequences from the RT-nested PCR, longer S segment sequences were obtained from two samples (V32/13, V36/13), whereas whole S and M segment and partial L segment sequences were obtained from the Haskovo isolate. All sequences have been submitted in GenBank DataBase and assigned the accession numbers KJ000206, KR011837–KR011839, KR092379, KR092381, KT334329, and KT334330. An additional S segment sequence obtained from a British traveler who returned to United Kingdom in June 2014 from a trip to a rural area of Burgas district near the Black Sea in Bulgaria (GenBank Acc. No KM201260) [Lumley et al., 2014] was included in the analysis. The phylogenetic trees based on sequences from S (whole ORF and partial), M (whole), and L (partial) RNA segments from Bulgarian CCHFV strains obtained from the present study and from the GenBank DataBase are shown in Figure 2.
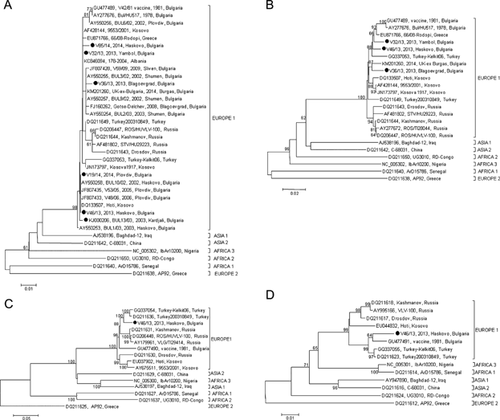
All CCHFV sequences from Bulgaria clustered in the “Europe 1” clade. The mean genetic difference of all available Bulgarian sequences in the S RNA segment (1374 nts) is 2.2% (range 0–2.8%). The sequence of the whole M RNA segment from the V46/13 isolate differs from the Bulgarian vaccine strain V42/81 by 6.2%; the respective difference in the partial L segment (465 nts) is 3.3%.
DISCUSSION
During 2013–2014 11 CCHF cases have been reported in Bulgaria, with seven of them occurring in the neighboring districts of Haskovo (n–= 4) and Kardzhali (n = 3). The human seroprevalence rates in these regions are 4.6% and 6% [Christova et al., 2013]. The highest seroprevalence (8%) has been reported in Burgas, where one case was reported in 2014 in a UK traveler [Lumley et al., 2014]. A recent study on livestock in farming communities of Burgas district revealed 71% seroprevalence in cattle, 74% in sheep and 60% in goats; remarkably, almost 50% of 1-year-old animals were found seropositive, while the respective rate among the 3-year-old animals is over 80% [Barthel et al., 2014]. Ten of the 11 patients of the study, as well as the British traveler, reported a tick bite. This finding is in accordance with the seroprevalence study which showed that the risk for seropositivity is increased 5.4-fold in persons bitten by ticks [Christova et al., 2013]. Furthermore, studies on ticks collected from livestock during 2006–2010 showed that 2–4.8% of ticks (mainly H. marginatum) in Kardzhali and Haskovo are CCHFV-infected [Gergova et al., 2012].
A relatively low genetic difference (2.2%) was seen among the S RNA segment sequences. The difference was higher in the M RNA segment (6.2%). The highest genetic diversity in the M RNA segment has been reported previously [Anagnostou and Papa, 2009]. This is not unexpected, since M segment encodes the two glycoproteins, Gn and Gc, which are involved in binding to the host cell receptors and may often accumulate mutations in order to become efficient for virus attachment and entry.
Complex and unresolved factors play a role in CCHFV epidemiology, evolution, and emergence in new foci. Understanding the CCHF epidemiology and virus evolution is the basis for public health programs and vaccine design. The present study provides an update into the geographic distribution and molecular epidemiology of CCHF in Bulgaria, suggesting that although the virus expands into new territories, it is more active in the established endemic foci which seem to offer the most suitable ecosystem and environment. The study showed that all CCHFV strains belong to Europe 1 lineage and present low genetic differences. Studies on ticks are needed to check whether the second European lineage (Europe 2) is present in Bulgaria, as is the case in other Balkan countries and Turkey.
ACKNOWLEDGMENTS
This work is part of the CCH Fever network (Collaborative Project, grant number 260427). We thank N. Kalvatchev, K. Tsergouli, and Elpida Gavana for excellent technical assistance.




