Identification and BAC construction of Han, the first characterized HCMV clinical strain in China
Abstract
Human cytomegalovirus (HCMV) is the leading infectious cause of birth defects, and may lead to severe or lethal diseases in immunocompromised individuals. Several HCMV strains have been identified and widely applied in research, but no isolate from China has been characterized. In the present study, we isolated, characterized and sequenced the first Chinese HCMV clinical strain Han, and constructed the novel and functional HCMV infectious clone Han-BAC-2311. HCMV Han was isolated from the urine sample of a Chinese infant with multiple developmental disorders. It expresses HCMV specific proteins and contains a representative HCMV genome with minor differences compared to other strains. By homologous recombination using mini-F derived BAC vector pUS-F6, the infectious clone Han-BAC-2311 was constructed containing representative viral genes across the HCMV genome. The insertion site and orientation of BAC sequence were confirmed by restriction enzyme digestion and Southern blotting. The reconstituted recombinant virus HanBAC-2311 expresses typical viral proteins with the same pattern as that of wild-type Han, and also displayed a similar growth kinetics to wild-type Han. The identification of the first clinical HCMV strain in China and the construction of its infectious clone will greatly facilitate the pathogenesis studies and vaccine development in China. J. Med. Virol. 88:859–870, 2016. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous pathogen, which can infect all the important organs, including the lungs, liver, kidney, blood, bone marrow, salivary gland, heart, and brain [Cobbs et al., 2002; Cheeran et al., 2009; Abdel-Latif Mel and Sugo, 2010]. The HCMV infection rate in industrialized countries such as USA is about 50–70%, but in developing countries such as China, the prevalence is over 90% [Fulop et al., 2013]. In spite of being harmless for healthy individuals [Mong et al., 1999], primary and recurrent HCMV infection causes severe or even lethal diseases in immune immature newborns and immunocompromised individuals, manifesting as pneumonitis, hepatitis, retinitis, and gastrointestinal diseases [Boppana et al., 2011; Mercorelli et al., 2011]. Congenital HCMV infection is the leading cause of birth defects, primarily presenting neurological sequelae in neonates [Stagno et al., 1980; Luo et al., 2010, 2008].
HCMV has a variety of different strains rather than defined virus genotypes which can display different virulence levels, tissue tropism, and pathogenicity [Kim et al., 2012]. Patients often acquire additional HCMV strain reinfections and suffer a mixed infection of HCMV strains and genotypes [Puchhammer-Stockl and Gorzer, 2011; Ross et al., 2011]. Even the standard HCMV strains used in the laboratory differ from their original clinical isolates both in biological and genomic properties [Murphy and Shenk, 2008]. AD169 [Chee et al., 1990; Murphy et al., 2003] and Towne [Dunn et al., 2003; Murphy et al., 2003], the two most broadly utilized laboratory strains, have been shown to have independently lost a multigene segment, 15 kb for AD169 and 13 kb for Towne. Toledo, a clinical strain, has an inverted 15 kb fragment and a disrupted UL128 ORF, which allows it to replicate more efficiently in fibroblasts [Cha et al., 1996]. Given this variation, it is of great importance to isolate and study additional clinical HCMV strains, to better understand viral pathogenesis and infection characteristics. Although there are many clinical and laboratory HCMV strains that have been used in various studies [Murphy and Shenk, 2008], none of them are from China. The lack of a native HCMV clinical strain limits pathology and epidemiology studies of HCMV infection in China, and development of a specific “Chinese” HCMV vaccine also requires access to locally isolated clinical strains.
Despite the observed variation between different HCMV strains, they share a genome approximately 230 kb in size which contains over 200 identified open reading frames (ORFs) [Dunn et al., 2003; Yu et al., 2003]. However, the large genome size raises challenges of manipulating and analyzing the genes with regular molecular cloning methods. To address this issue, the bacterial artificial chromosome (BAC) [Sternberg, 1990; Shizuya et al., 1992] has been deployed as a cloning vehicle in HCMV [Marchini et al., 2001] and other herpes viruses [Marchini et al., 2001; Zhang et al., 2007] since 1997, when Messerle reported the first infectious herpes virus clone using BAC [Messerle et al., 1997]. As a well-established low-copy replicon unit derived from the bacterial fertility plasmid (F factor), the BAC can accommodate large segments of genomic DNA and permits further modification in E. Coli, which provides an ideal platform for HCMV study, especially HCMV genomic modification and elucidation of viral gene function, and helps retain the characteristics of the original virus isolate. In previous studies, F factor derived pMBO1374 was further modified to obtain pUS-F3 and pUS-F5, which was used to construct the infectious clones of HCMV Towne strain and varicella-zoster virus (VZV) Oka strain, respectively [Marchini et al., 2001; Zhang et al., 2007].
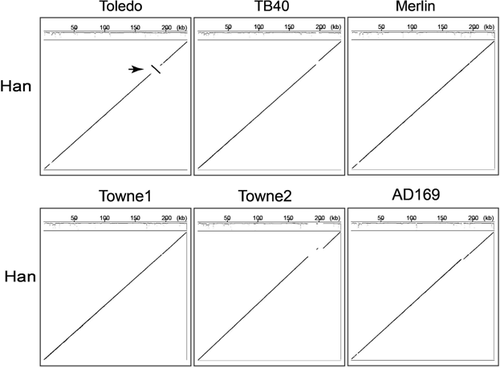
In the present study, we report the first identified HCMV clinical strain Han, which was isolated in China from a native patient with maldevelopment of multiple organs. The genome of the virus strain was sequenced and compared with those of Toledo, TB40, Merlin, Towne, and AD169 strains. The analysis indicated that the Han strain retains the standard overall genome structure, but contains an ∼15 kb inversion with respect to the Toledo strain, consistent with the other strains investigated here. HCMV Han-BAC was constructed using pUS-F6, a BAC vector derived from pUS-F5. The generated infectious clone, Han-BAC-2311, contains the wild-type HCMV Han genome with a 5.5 kb deletion of US2–US9, without gene rearrangement around the BAC insertion site. The reconstituted virus, HanBAC-2311, expresses typical HCMV proteins of all three phases, and displays similar growth kinetics compared with the Han wild-type virus. The isolation and characterization of the first HCMV clinical strain in China will be valuable for studying the pathology and epidemiology of HCMV in China, and provides a potential candidate suitable for HCMV vaccine development in China. The construction of Han-BAC establishes a solid platform for further study of gene functions and pathogenesis, facilitating Han derived vaccine development and screening of anti-viral reagents.
MATERIALS AND METHODS
Patient Description
A female baby was born to a primigravida mother at term with a birth weight of 3.0 kg, and had been breast-fed. At 5-months old, the patient developed progressive jaundice and was admitted to the hospital with a weight of 5.6 kg. The skin and sclera manifested obvious yellow discoloration. Physical examination showed signs of hepatosplenomegaly (liver span 6 cm below the right sub-costal margin, and spleen 4 cm below the left sub-costal margin). Liver functional tests showed dramatically elevated levels of alanine aminotransferase (ALT, 380 U/L, reference 0–40 U/L), aspartate transaminase (AST, 1047 U/L, reference 5–34 U/L), total bilirubin (TBil, 488 µmol/L, reference 3.4–20.5 µmol/L), and direct bilirubin (DBil, 400 µmol/L, reference 0–8.6 µmol/L). Further examination demonstrated congenital heart disease (CHD) with coarctation of the aorta, patent ductus arteriosus and atrial septal defect. The laboratory test for HCMV infection was positive (IgM 1:32, viral antigen pp65 is positive), and the viral DNA load in urine was 1.59×104 copies/ml by quantitative PCR (qPCR).
Specimen Collection and Virus Isolation
The urine specimen of the patient was collected and treated with an antibiotic cocktail containing 500 U/ml penicillin, 500 µg/ml streptomycin and 500 µg/ml kanamycin. After centrifugation at 300g for 15 min, the supernatant was applied onto a human embryonic lung fibroblasts (HELs) monolayer, and incubated at 37°C overnight to allow virus adsorption. Then the inoculum was removed and cells were washed with Hank's buffer. The inoculated HELs were maintained in Modified Eagle's medium (MEM) supplemented with 2% fetal bovine serum (FBS), penicillin (100 units/ml)/streptomycin (100 μg/ml) and 2 mM l-glutamine (Gibco/Life Technology). Culture medium was refreshed every 5 days and cells were passaged when necessary. The cultures were observed every other day to monitor appearance of cytopathic effects (CPEs). When HCMV characteristic CPEs appeared, the culture was cultured for an additional three days and then passaged. To prepare cell free viruses, supernatant and the cells were harvested separately when specific CPEs appeared in 90% of cells. The harvested cells were freeze-thawed three times to increase the virus release, and then resuspended in the harvested supernatant. Cell debris was removed by centrifugation twice at 400g for 10 min. Then the supernatant was aliquoted and stored at −80°C as virus stock for further experiments. This HCMV clinical isolate was completely sequenced, and named as Han according to the patient's family name.
Sequencing and Genome Comparison
The genome of HCMV Han was sequenced by BGI on an Illumina HiSeq (Shenzhen), and the complete sequence is available under the GenBank accession number KJ426589.1. Total reads were ∼6 × 106 yielding ∼2.1 × 103 coverage. All available HCMV genomes as of 23/10/2014 were downloaded from GenBank. Of these, Han (KJ426589.1), Toledo (GU937742.1), TB40 (KF297339), AD169 (FJ527563.1), Towne1 (FJ616285.1), Towne2 (AY315197.2), and Merlin (AY446894.2) were selected for comparative analysis. Two Towne strains were selected since a preliminary pairwise nucleotide alignment indicated they were sufficiently distinct.
The Mugsy multiple genome alignment tool [Angiuoli and Salzberg, 2011] was used to perform pairwise comparison of each genome pair and to identify rearrangements and deletions/insertions. A graphical representation of the multiple alignment format (MAF) that was output for each comparison was visualized using Gmaj [Blanchette et al., 2004] and was also parsed using an in-house script (available from the authors on request) to identify the major evolutionary events (i.e., deletion/insertion and rearrangements) and provide a text summary of the graphical representation. In addition, the Artemis Comparison Tool was also applied to perform pairwise comparison of genomes and to further investigate specific regions of interest.
Generation of HCMV Han-BAC Clones
The mini F plasmid pUS-F6 was derived from pMBO1374 [Oconnor et al., 1989]. Both left and right flanking sequences for homologous recombination in eukaryotic cells were amplified using viral DNA of HCMV Han as the template. The primers for flankings are listed in Table I.
| Forward primer (5′–3′) | ||
|---|---|---|
| Targets | Reverse primer (5′–3′) | Product sizes (bp) |
| Left flanking | CGGGATCCGGGCAGTGGGAGTTCATGTT | |
| 1,601 | ||
| CCCAAGCTTAGCGAGAGCACTGGCAGGGG | ||
| Right flanking | CCCAAGCTTGAGGGTACTGGGGCAGACGG | |
| 2,000 | ||
| CGGGATCCGTCCCCCGCACCCTAAAACA | ||
| UL31 | CGGGATCCAACGACGGCGGCGGCGACCAGATTATG | |
| 1,788 | ||
| CCCTCGAGGATGGGGAGACGTGAGAAAGTTGTCCGCG | ||
| UL33 | AAGGACGACGATGATAAGATGGACACCATCATCCAC | |
| 1,357 | ||
| CGGAATTCGTGTAGGAAGCTGTTCTGCAGGTAGTGGCG | ||
| UL34 | ATGAACTTCATCATCACCAC | |
| 1,224 | ||
| TTAAATACACAACGGGGTTATGGG | ||
| UL37 | CCGCTCGAGATGTCTCCAGTCTACGTGAATCTGTTAG | |
| 2,798 | ||
| GGGGTACCTCAGACGATCCGATGAACGTCG | ||
| UL44 | CCCAAGCTTATGGATCGCAAGACGCGCCTC | |
| 1,302 | ||
| CGCGGATCCCTAGCCGCACTTTTGCTTCTT | ||
| UL69 | CCCAAGCTTATGGAGCTGCACTCACGCGGC | |
| 2,229 | ||
| CCGGAATTCTTAGTCATCCATATCATCGCT | ||
| UL82 | ATGTCTCAGGCATCGTCCTC | |
| 1,680 | ||
| CTAGATGCGGGGTCGACTGC | ||
| UL97 | CCCAAGCTTATGTCCTCCGCACTTCGGTCT | |
| 2,124 | ||
| CCGGAATTCTTACTCGGGGAACAGTTGGCG | ||
| IE1 | CATATGATGGAGTCCTCTGCCAAGAGAAAG | |
| 1,758 | ||
| CGCGGATCCTTACTGGTCAGCCTTGCTTCTAGT | ||
| IE2 | CGGAATTCATGGAGTCCTCTGCCAAGAGA | |
| 3,397 | ||
| CGGGATCCTTACTGAGGAACAAGTCTCAGTAA | ||
| gB | CGCGGATCCATGGAATCCAGGATCTGGTG | |
| 2,700 | ||
| CCCAAGCTTTCGGACGTTCTCTTCTTCGT | ||
| US29 | CTAGACTAGTGCTCTACAGTGGGTGGTGGT | |
| 500 | ||
| CCCAAGCTTAAGCGTTGCCGTAGCTGGCG |
- Restriction enzyme sites are underlined.
The PCR products of both flanking sequences were cloned into the F-plasmid vector pUS-F5 [Zhang et al., 2007] to generate pUS-F6. pUS-F6 was linearized by BamHI digestion and transfected into wild-type HCMV Han infected HELs by electroporation. The cells were then maintained under standard cell culture conditions until CPEs and eGFP signal were clearly apparent. The plaques with eGFP+ cells were picked up, and then inoculated to fresh HELs for amplification and purification of the recombinant virus.
The purified eGFP+ recombinant virus was applied to infect serum-starved HELs. When the infected cell started to express eGFP, cells were harvested and circular HCMV Han-BAC DNA was isolated from these cells as described elsewhere [Yu et al., 2002]. After brief washing, cells were resuspended in solution I (10 mM Tris pH 8.0, 10 mM EDTA), and lysed by supplement proteinase K (0.25 mg/ml), sodium dodecyl sulfate (SDS, 0.6%) and NaCl (1 M). After 2 hr incubation at 50°C, cell debris was clarified by centrifugation. The supernatant was processed with phenol-chloroform extraction assay twice, followed by ethanol precipitation, and DNA was dissolved in ddH2O. Then the HCMV Han-BAC DNA was transformed into E. coli DH10B competent cells (Invitrogen) by electroporation using a Gene Pulser (Bio-Rad) with a 1-mm-gap cuvette at 1.6 kV, and plated on Luria-Bertani (LB) agar containing 25 µg/ml chloramphenicol for selection.
Identification of Han-BAC-2311 Clone
HCMV Han-BAC clones were first screened by colony PCR, and the Han-BAC-2311 clone was selected for further identification of the representative HCMV genes. Wild-type HCMV Han genomic DNA served as a positive control. The primers used for identification of Han-BAC-2311 clone by PCR are listed in Table I.
The insert position and direction of BAC sequence was further confirmed by Southern blotting. Left and right flanking sequences were labeled with DIG-dUTP using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche), and used as probes on BamHI or HindIII digested Han-BAC-2311 DNA.
Rescue Viruses From Han-BAC-2311
DNA was isolated from the culture of Han-BAC-2311 monoclone using a NucleoBond Xtra Midi kit (Macherey-Nagel, Germany) following the manufacturer's protocol. The Han-BAC-2311 DNA (8 µg) was transfected into 8×106 HELs by electroporation using a Gene Pulser with 4-mm-gap cuvettes at 260 V/75 Ω/975 µf. To facilitate virus growth, 4 µg of pcDNA3-pp71 was co-transfected into HELs [Baldick et al., 1997]. The cells were then maintained under standard cell culture conditions until CPEs and eGFP signal were clearly apparent. The rescued virus was named Han BAC-2311, and the virus stock was prepared as described previously [Pan et al., 2013].
Immunoflorescent Assay (IFA), Western Blotting
IFA and western blotting was performed as described previously [Duan et al., 2014]. The following mouse monoclonal antibodies were applied: anti-IE1 (IgG2a), -gB (IgG2b, provided by Dr. William Britt at University of Alabama), -IE1/2 (IgG1), -UL44 (IgG1), -pp65 (IgG1, Virusys), -β-actin (IgG1, Santa Cruz Biotechnology), and the corresponding secondary antibodies including Alexa Fluor 488 (AF488) conjugated goat anti-mouse IgG2a, AF488 conjugated goat anti-mouse IgG2b (Invitrogen), R-PE conjugated goat anti-mouse IgG1 (Southern Biotech), and Horseradish Peroxidase (HRP) conjugated goat anti-mouse IgG (Abbkine).
Quantitative Real-Time PCR (qPCR)
To compare viral growth kinetics, HELs were infected with the indicated viruses at an MOI of 1. Total DNA was extracted from the cultures for qPCR. Copy number of UL83 gene of the viral genome was determined according to the standard curve generated by using pCDNA3-UL83 plasmid with primers UL83-F 5′- GCGAGACCGTGGAACTGC -3′ and UL83-R 5′- TTGCCCTGGATGCGATACTG -3′. In Han genome, the primers amplifies the region from nucleotide 121,544 to 121,676. The results were taken from three independent experiments and presented as mean ± SD.
RESULTS
Identification of the Isolate HCMV Clinical Strain
After inoculation with the patient's urine specimen, the inoculum was removed and replaced with fresh media. HELs were cultured and passaged until specific CPEs appeared at 8 weeks post inoculation. One week later, the cells were added onto naïve HELs monolayers, and cells and supernatant were both collected when CPEs were observed in 90% of the cells. Virus stocks were prepared and titrated for further identification, including viral genome analysis by sequencing (data not shown) and viral gene products by IFA.
Virus stock was applied to infect HELs at an MOI of 1. Compared to the mock-infected controls, virus infected HELs at 72 hpi lost the typical long, thin and bi-polar fibroblast morphology, and turned into enlarged round cells with decreased attachment to the culture surface (Fig. 1A). Representative viral products of all the three phases of immediate early (IE) protein IE1, early protein UL44 and pp65, and late protein glycoprotein B (gB) were expressed in infected cells. At 72 hpi, IE1 and UL44 were located in the nuclei, whereas pp65 and gB were distributed in both the nuclei and cytoplasm, but were mainly located in the cytoplasm at this time point (Fig. 1B). These results are consistent with observations for standard HCMV strains, and confirmed the virus isolate as HCMV.

Comparison of HCMV Han Genome With Other Strains
The sequence of HCMV Han was compared with other representative HCMV clinical strains including TB40, Toledo and Merlin. Towne and AD169 were included as they represent standard laboratory strains. The overall GC content of HCMV Han is 57.5%, consistent with values reported for other strains. Pairwise comparisons of all combinations of the six genomes were considered in this study and are summarized in Figure 2. At the genome level, the overall genome structure is consistent with these five reference strains and divided into six regions, TRL, UL, IRL, IRS, US, and TRS. Compared to the Toledo strain, the Han has an inversion of ∼15 kb from 176,729 to 191,263 bp, corresponding to genes UL128 and UL129 at one end and UL133–UL148 at the other, and consistent with previous reports [Murphy et al., 2003].
Creation of HCMV Han-BAC Clones
During continuous culture and propagation, HCMV AD169 lost a 15 kb fragment as well as the capability to propagate in vivo, which also changed the capacity for pathogenesis [Cha et al., 1996]. To maintain the integrity of the first characterized HCMV clinical isolate in China, we attempted to construct a BAC containing as much of the intact HCMV Han genome as possible. Since the US region of HCMV genome has been demonstrated to be dispensable for viral replication in most cell types [Jones and Muzithras, 1992; Dunn et al., 2003], this segment was used as the insertion site for the Han-BAC construction. The flanking fragments (1.6 kb left arm and 2.0 kb right arm) were synthesized by PCR with the primers listed in Table I, which contain HindIII and BamHI sites at the ends. These PCR products were digested with BamHI and ligated to each other, resulting in a 3.6 kb linear fragment which was then digested with HindIII and cloned into the unique HindIII site in pUS-F5 to create BAC vector pUS-F6 (Fig. 3A). The BAC vector pUS-F6 contained the following critical components: (i) sequences matching the viral genome to allow homologous recombination; (ii) prokaryotic genetic elements necessary for maintenance in bacteria; (iii) chloramphenicol resistant marker for prokaryotic selection; and (iv) the enhanced green florescent protein (eGFP) reporter driven by the SV40 promoter for further identification and purification of Han-BAC reconstituted virus in eukaryotic cells.
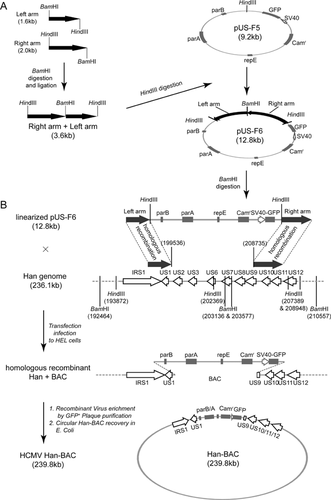
BAC vector pUS-F6 was linearized by BamHI digestion and introduced into HELs infected with wild-type HCMV Han. Homologous recombination between pUS-F6 and wild-type HCMV Han genome occurred during viral replication in Han infected HELs (Fig. 3B).
Confirmation of HCMV Han-BAC Monoclone
PCR was applied to amplify various regions of the viral genome to identify the clones. Representative viral genes distributed across the HCMV genome were examined, including UL31, UL33, UL34, UL37, UL44, UL69, UL82, UL97, IE1, IE2, gB, and US29. A candidate infectious monoclonal Han-BAC-2311 was identified to express all the examined genes (Fig. 4A). For the restriction fragment length polymorphism analysis (RFLP), HCMV Han viral genome DNA and Han-BAC-2311 DNA were digested by BamHI or HindIII, and separated by 1.0% agarose gel. Most of the digestion fragments of Han-BAC-2311 were similar Compared to the digestion result of Han with only two main visible differences, an lost 5.0 kb fragment by HindIII digestion and an gained 21.8 kb fragment by BamHI digestion (Fig. 4B left panel), which is consistent with the simulated digestion maps generated by SnapGene (Fig. 4B right panel). Besides the two clearly distinguishable differences, the recombination also changed other fragments, but which were hard to be identified because of the similar sizes with the non-chanced fragment (data not shown). To further confirm that Han-BAC-2311 was the positive clone, Southern blotting was perform by detecting the left and right flanking areas. As shown in Figure 3, the successful homologous recombination deleted the 5.5 kb fragment from US2 through US9 in the Han genome, and replaced it with the 9 kb fragment of the pUS-F6 sequence. Southern blotting on Han-BAC-2311 presented the same pattern as expected. For wild-type HCMV Han, HindIII digestion showed 8.4 and 5.02 kb fragments when hybridized with left and right arm probes respectively. The fragment of the 5.5 kb US region containing one HindIII digestion site was replaced with the 9 kb BAC containing two HindIII sites in the flanking arms at each ends, which reduced the detected fragments to 5.6 kb (left arm probe) and 2.35 kb (right arm probe) for Han-BAC-2311. When digested with BamHI, wild-type HCMV Han DNA showed 10.4 kb and 6.9 kb fragments, while Han-BAC-2311 DNA yielded two 21.6 kb fragments with left and right arms, respectively (Fig. 4C). The Southern blotting data was consistent with the expected pattern and indicates the BAC sequence is located in the designed site and no genome rearrangement occurred around this region.
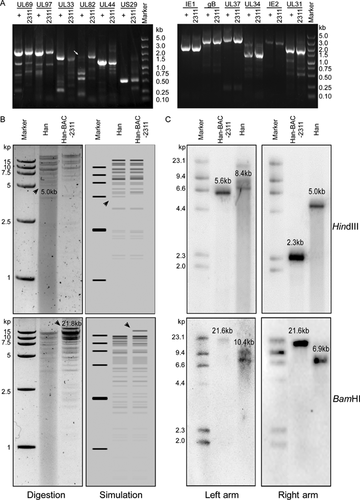
Taken together, the data indicates that the HCMV infectious monoclone Han-BAC-2311 matched the predicted structure of the recombinant HCMV Han-BAC with homologous recombination between HCMV Han genome and pUS-F6.
The Reconstituted Virus HanBAC-2311 Shares Similar Features With Wild-Type Strain
Finally, the recombinant virus was rescued from Han-BAC-2311 and named HanBAC-2311. The HELs electroporated with Han-BAC-2311 DNA clearly showed eGFP signal (Fig. 5A). HanBAC-2311 generated from the single GFP+ plaque induced typical HCMV CPEs and strong eGFP signal at 7 dpi (Fig. 5B).
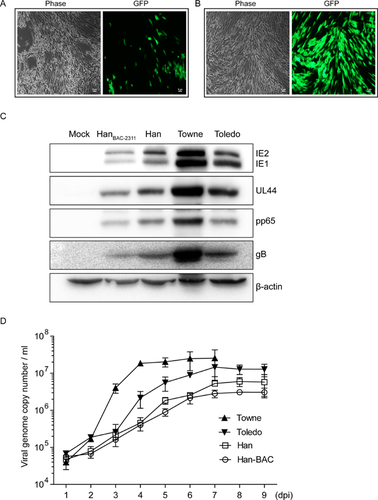
HanBAC-2311 was then examined for fundamental biological characteristics. Western blot analysis revealed that HanBAC-2311 infected HELs expressed the representative viral proteins with similar levels to wild-type HCMV Han despite a slight abatement (Fig. 5C). The viral replication kinetics was in accordance with the protein expression levels (Fig. 5A). HanBAC-2311 and Han exhibited very similar growth kinetics, and both were lower than those of Towne and Toledo. The viral protein synthesis and growth curve were consistent with the appearance of CPEs, which were visible in both HCMV Han and HanBAC-2311 infected HELs at 6–7 dpi, later than that of Towne (3 dpi) and Toledo (4–5 dpi) (data not shown).
Collectively, HanBAC-2311 retains the essential properties of wild-type HCMV Han, aside from minimal phenotypic restraint in replication kinetics, and Han-BAC-2311 can be considered as an infectious BAC clone of the first isolated clinical HCMV strain Han in China.
DISCUSSION
In spite of being harmless for healthy adults, the congenital, perinatal, and postnatal HCMV infection, may lead to presentation of severe syndromes in newborns. In particular, congenital HCMV infection is a leading cause of disabilities in newborns, including low birth weight, microencephaly, encephalitis, seizures, cerebral palsy, petechial rash, and moderate hepatosplenomegaly with jaundice [Revello et al., 2006; Luo et al., 2010; Karltorp et al., 2012]. Epidemiology studies reveal over 1–3% of newborns are congenitally infected by HCMV in China [Zhong and Ma, 1993; Wen et al., 1996], which is higher than the prevalence in the United States. Considering 10–15% of the congenital HCMV infected infants show clinical abnormalities at birth (symptomatic infection), nearly 40,000 Chinese neonates per year suffer sequelae caused by congenital infection. Only limited, and more commonly, lack of, systematic clinical data are available on newborns with symptomatic congenital HCMV infection. The HCMV screening system is neither well developed nor well performed and is normally overlooked during pregnancy or after birth. The extremely high HCMV infection prevalence (over 90%) represents a major transmission source. Newborns or infants are more readily infected due to their immature immune system, which leads to severe diseases such as pneumonia, hepatitis, hepatosplenomegaly, thrombocytopenia, and atypical lymphocytosis.
In this study, the 5-month old patient displayed a series of clinical symptoms including signs of hepatitis with jaundice, hepatosplenomegaly, and other maldevelopments. Evidence from laboratory tests indicated that the symptoms were associated with HCMV infection, but there was no direct evidence to indicate whether the infection occurred congenitally, perinatally, or postnatally. Congenital biliary atresia was shown, which is possibly induced by HCMV, and likely caused by congenital and postnatal infection at early childhood [Xu et al., 2012]. This patient also had CHD with aortic stenosis, patent ductus arteriosus and atrial septal defect, but there is currently no data supporting an association between CHD and a congenital HCMV infection, even though such an infection might cause multiple system malformation.
Various HCMV strains have been isolated and used in basic and clinical research. So far, GenBank contains 52 HCMV strains or isolates with entire genome sequences. However, HCMV Han is the only GenBank listed clinical strain that originates from China. To understand the specific pathogenesis in HCMV infected patients in China, it is necessary to access native HCMV clinical strains. In this study, HCMV Han was isolated from a native 5-month patient with developmental disorders. The developmental disorders may be related to congenital HCMV infection, but this cannot be confirmed because of the time of diagnosis.
The variation among HCMV strains is possibly related to variation in cell tropism, immunopathogenesis, virulence, or immune evasion [Hahn et al., 2004]. For example, laboratory strain AD169 (GenBank X17403), with attenuated virulence and different tropism, lacks the 15 kb segment in the UL/b′ region found in some low passaged clinical strains including Toledo [Cha et al., 1996]. Although the exact function of the 19 open reading frames (ORF) in this 15 kb segment is still not fully clarified, they appear to play a crucial role in pathogenesis [Wang et al., 2005]. Other strains with different genome sequences similarly display different infection characteristics to varying degrees. In this study we have only performed a relatively superficial investigation of the genome sequence with the primary goal of confirming the overall genome structure is retained in this strain. A more detailed investigation of the specific differences are beyond the scope of this report and will be presented in the future.
Viral BACs serve as a basic platform for detailed studies on viral gene function and pathogenesis and provide a new strategy for vaccine development. Manipulation of the viral genome, such as deleting negative immune regulators or inserting immunogenic target elements, can enhance the safety and efficacy profiles of vaccine candidates. Many studies have exploited the BAC to develop large antigen-capacity DNA vaccines (BAC-VAC) against complex pathogens, such as herpes simplex virus 1 (HSV-1) [Suter et al., 1999], herpes simplex virus 2 (HSV-2) [Meseda et al., 2004], Epstein-Barr virus [Delecluse et al., 1998], and pseudorabies virus [Smith and Enquist, 1999]. Moreover, the virus-BAC is also an effective way to keep the original entire viral genome intact to avoid accumulation of mutations during long-term culture or passaging. Here, HCMV Han-BAC-2311 was generated by introducing the BAC sequences derived from pUS-F6 into the HCMV Han genome. The replication kinetics of the virus rescued from Han-BAC-2311 is similar to the wild-type HCMV Han.
In conclusion, we have isolated the Han strain, the first human cytomegalovirus clinical strain from China, and generated a virus-BAC system to contain the virus genome. Experimental results indicate the virus-BAC has similar characteristics to the wild-type and thus represents an important vehicle for studying HCMV epidemiology and furthering vaccine development efforts targeting HCMV strains in the country. Additional experimental studies and bioinformatics analysis will allow us to gain further insights into the genome features that characterize the various strains that have been isolated.
AUTHORS’ CONTRIBUTION
FZ, ZZS, and ZYL contributed equally to this work. QR and MHL designed and instructed the study. ZYL and YPM collected the clinical information and isolated the virus. FZ, ZZS, and ZYL constructed and identified the Han-BAC. WBZ and SC contributed in western blot and IFA. SR performed the bioinformatics analysis. FQX and HZ offered valuable comments and suggestions for BAC construction and identification. BY, GHQ, HFJ, SG provided experimental support. FZ and ZZS drafted the manuscript. All authors have read and approved the final manuscript for submission.
ACKNOWLEDGMENT
We thank Dr. William Britt (University of Alabama, USA) for providing HCMV specific antibodies.




