The clinical implication of single nucleotide polymorphisms in deoxycytidine kinase in chronic hepatitis B patients treated with lamivudine
Abstract
Deoxycytidine kinase (dCK) is a critical enzyme involved in intracellular phosphorylation of lamivudine (LAM) to its active triphosphates. We conducted this study to determine dCK polymorphisms in Koreans and to evaluate whether the discovered single nucleotide polymorphisms (SNPs) were associated with treatment outcomes in chronic hepatitis B (CHB) patients treated with LAM. The full-length dCK gene was sequenced from 24 healthy volunteers and 24 patients with CHB. One hundred twenty-seven patients with CHB who were followed-up for at least 24 months after LAM treatment were enrolled. Virological response as determined by undetectable HBV DNA was defined as a good drug response. Primary non-response at 6 months and virological breakthrough within 12 months were defined as a poor drug response. Six novel dCK SNPs were found (−2052C/A, IVS3 − 46G/del, IVS4 + 40G/T, IVS5 + 39T/C, IVS5 − 72A/T, and 966–975T10/T11). In particular, two promoter SNPs, namely −360C/G and −201C/T, were in full linkage disequilibrium. These two SNPs had a higher allele frequency than previously reported in Caucasian, Japanese, and Chinese (26% vs. 2%, 13.1%, and 15.6%, respectively). There was no significant difference between treatment response groups in terms of the distributions of SNP genotypes or allele frequencies. However, there was significant difference in the allele frequency of −360G/−201T between HBeAg seroclearance group and HBeAg non-seroclearance group (P = 0.045). In conclusion, six novel dCK SNPs were discovered. Two promoter SNPs, namely −360C/G and −201C/T, were more frequent in Koreans than other populations. In particular, HBeAg-positive patients with the −360G/−201T haplotype may help HBeAg seroclearance in response to LAM therapy. J. Med. Virol. 88:820–827, 2016. © 2015 Wiley Periodicals, Inc.
Abbreviations
-
- HBV
-
- hepatitis B virus
-
- dCK
-
- deoxycytidine kinase
-
- LAM
-
- lamivudine
-
- SNPs
-
- single nucleotide polymorphisms
-
- CHB
-
- chronic hepatitis B
-
- NAs
-
- nucleoside/tide analogues
-
- NRTIs
-
- nucleoside-based reverse transcriptase inhibitors
-
- HBeAg
-
- hepatitis B e antigen
-
- Anti-HBe
-
- hepatitis B e antibody
-
- ALT
-
- alanine aminotransferase
-
- HIV
-
- human immunodeficiency virus
-
- HCV
-
- hepatitis C virus
-
- HDV
-
- hepatitis D virus
-
- ULN
-
- upper limit of normal
INTRODUCTION
Potent anti-viral agents have been developed to treat chronic hepatitis B virus (HBV) infection. With the development of the anti-viral agents, antiviral resistance has become an important problem during long-term antiviral therapy. Recently, HBV variants with multi-drug resistance has been unresolved issue in patients who received sequential treatment with nucleos(t) ide analog (NA) [Villet et al., 2006; Yim et al., 2006; Lee et al., 2014]. NAs must be given for several years in most cases to achieve sustained viral suppression. Unfortunately, the durability of viral suppression is not continued after cessation of NAs. Therefore, a long duration of NA treatment is associated with an increased risk of emergence of drug resistance [Perrillo, 2004; Kim do et al., 2013; Ze et al., 2014].
Currently, LAM is still recommended as a first-line therapy in the APASL guidelines because of its low cost, and safety despite the low potency and low genetic barrier to resistance [Liaw et al., 2008]. In terms of treatment failure, viral resistance to antiviral agents and poor compliance are the most important factors. Interestingly, another important point is that host factors can affect anti-viral efficacy. However, it remains unclear if any host factors play a role in the response to anti-HBV drugs [Locarnini and Mason, 2006].
Recent genetic studies reported single nucleotide polymorphisms (SNPs) in HLA-DPA1 and HLA-DPB1 genes are associated with risk of persistent infection of HBV [Kamatani et al., 2009]. In addition, SNPs in genes encoding target enzymes have an important role as predictive factors for chemotherapeutic drug response [Evans and Relling, 1999; Hoehe et al., 2003]. Especially, favorable SNPs in the IL28B gene are associated with treatment efficacy of pegylated interferon and spontaneous viral clearance in patients with HCV [Ge et al., 2009]. However, there were few data on the association between genetic background and response to antiviral agents of HBV. To date, SNPs’ application is not recommended for predicting treatment outcomes in patients with chronic hepatitis B (CHB) [Stattermayer et al., 2014].
NAs are prodrugs that must be metabolized intracellularly to gain activity. Three different kinases, deoxycytidine kinase (dCK), thymidine kinase, and deoxyguanosine kinase, have been found to be involved in LAM phosphorylation. Among these, dCK is considered as a critical enzyme because phosphorylation of dCK into its monophosphate form is the initiating step for activation of LAM [Kim and Ives, 1989; Shewach et al., 1992]. The dCK gene resides on the short arm of chromosome 4(4q13.3–q21.1). It has seven exons and extends over 34 kb [Chen et al., 1995; Johansson et al., 2000; Baker et al., 2013]. Interestingly, SNPs of dCK gene were reported as genetic markers for prediction of chemotherapy responsiveness in acute myeloid leukemia patients [Veuger et al., 2000; Shi et al., 2004]. However, no prior study has investigated whether dCK contributes to the responsiveness of LAM. We therefore investigated the potential influence of genetic variants of the dCK gene on clinical sensitivity to LAM. Genotyping of dCK candidate SNPs and their association with antiviral efficacy will be helpful in optimizing personalized drug therapy.
Our aims were to confirm known dCK polymorphisms in Chinese, Japanese, and Caucasian subjects, to discover new dCK polymorphisms in Korean patients, and to evaluate whether the discovered SNPs were associated with treatment outcomes of LAM therapy in patients with CHB.
PATIENTS AND METHODS
Study Population
A total of 127 patients with chronic hepatitis B who visited Severance Hospital between January 2003 and December 2006 were enrolled in this study. They were followed-up for at least 24 months after LAM treatment. Patients were eligible if they met the following entry criteria: they were 18–75 years of age; the presence of serum HBsAg was observed for at least 6 months; they had an elevated serum alanine aminotransferase (ALT) level on two occasions, at least 1 month apart, with an average value of ≥2 times the upper limit of normal (ULN); the presence of serum HBV DNA was documented on two occasions, at least 1 month apart. Additional requirements included a hemoglobin value of ≥10 g/dl, a platelet count of ≥100,000 mm3, a white cell count of ≥4,000 mm3, a polymorphonuclear count of ≥1,500 mm3, and normal renal function with normal serum creatinine levels. Candidates were required to have compensated liver disease with a prothrombin time of less than 4 sec, prolonged over control values, a serum albumin of ≥3.0 g/dl, a total bilirubin of ≤4 mg/dl, and no history of hepatic encephalopathy or bleeding esophageal varices.
Exclusion criteria were as follows: a history of corticosteroid treatment within 6 months of entry; previous therapy with IFN; the presence of antibodies to human immunodeficiency virus (HIV), hepatitis C virus (HCV), or hepatitis D virus (HDV); a history of malignancy or evidence of other forms of liver disease; a history of intravenous drug abuse. Patients with other significant medical or psychiatric problems were also excluded.
Patients were treated with LAM monotherapy for virological response. LAM was given at a dose of 100 mg per day for at least 12 months. Serum HBeAg, anti-HBe, HBV DNA, and ALT were assessed every 3 or 6 months, and whenever necessary during medication and after drug cessation. In addition, peripheral blood samples from 24 healthy volunteers and 24 patients with CHB were used as control samples. To overcome selection bias, healthy and CHB volunteers were matched for age and sex by simple random sampling (SAS 9.13 software). Two groups did not show any differences in the age and sex. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Board of Severance Hospital, and written informed consent was obtained from each patient.
Definitions
Patients without a decline in serum HBV DNA by ≥1 log10 IU/ml after the first 6 months of therapy were classified as “primary non-response (NR).” Virological breakthrough (VB) was defined as patients with an increase in serum HBV DNA of >1 log10 IU/ml (10-fold) above nadir after achieving virological response during continued treatment [Lok et al., 2007]. HBeAg seroclearance was defined as loss of detectable levels of HBeAg and HBV DNA in serum.
Patients who achieved virological response with undetectable HBV DNA for at least 48 weeks were classified as good drug responders. In contrast, patients with NR or VB were considered poor drug responders.
Assay Methodology
Commercially available enzyme-linked immunoadsorbent assays (ELISAs) were used for the detection of serum HBsAg, anti-HBs, anti-HBc, HBeAg, and anti-HBe (Abbott Laboratories, North Chicago, IL). Serum HBV DNA concentration was determined by a commercially available quantitative assay (Lightcycler 2.0, Roche Diagnostics, Branchburg, NJ). The sensitivity of the assay is approximately 300 copies/ml.
Sequence Analysis of the dCK Gene
Peripheral blood (10 ml) was collected from each individual. Genomic DNA from peripheral blood stored samples was isolated using a NucleoGen genomic DNA isolation kit (NucleoGen, Seoul, Korea) according to the manufacturer's instructions, and extracted DNA was subsequently stored at 4°C until analysis. Thirteen primer sets were used to amplify all seven DCK exons and the core promoter region for a total sequence length of 2862 bp (Supplemental Table I). Several fragments of the dCK gene were amplified with forward primers and reverse primers in a final volume of 25 μl containing 50–100 ng of genomic DNA, 20 pmol of each primer, 0.2 mM dNTPs, 2 U of Taq polymerase (iNtRON Biotechnology, Seoul, Korea) and manufacturer-supplied polymerase chain reaction (PCR) buffer. Amplification was performed in a GeneAmp PCR system 9700 (Perkin Elmer Corp., Norwalk, CT). PCR conditions were 94°C for 5 min, followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 50 sec, with final extension at 72°C for 7 min. PCR products were identified by electrophoresis, then purified with a PCR purification kit (iNtRON biotechnology, Seoul, Korea) and analyzed by direct sequencing (Bionex Co. Ltd, Seoul, Korea). Twenty-four healthy volunteers and 24 patients with CHB were screened for SNPs in the dCK gene.
Statistical Analysis
All data are expressed as means ± standard deviations (SDs). Student's t-test was used to compare mean age, pretreatment ALT level, and HBV DNA level among subject groups. χ2 test or Fisher's exact test was used to compare the sex ratio and frequencies of novel and known dCK genotypes. Based on gene frequencies, predicted phenotype frequencies were calculated according to the Hardy–Weinberg equation and compared with the observed frequencies using the χ2 test. Multivariate analyses were performed using logistic regression to identify factors associated with good response, and HBeAg seroclearance. Cumulative HBeAg seroclearance rates were estimated using the Kaplan–Meier method and compared by log-rank test. All statistical analyses were performed using the Statistical Package for Social Science (SPSS Inc., Chicago, IL) version 12.0. A P value of less than 0.05 was considered significant.
RESULTS
SNPs in the dCK Gene in Korean Subjects
All 5480 bp of the dCK gene were analyzed in a group of 24 healthy volunteers and 24 patients with CHB. Six SNPs were detected in the 5′ regulatory region. They were −2052C/A, −1431A/G, −1359A/G, −1329C/T, −360C/G, and −201C/T with allele frequencies of 0.979:0.021, 1:0, 1:0, 0.979:0.021, 0.74:0.26, and 0.734:0.266, respectively. We also detected 10 SNPs, 12 SNPs, and 1 SNP in introns, exons, and the 3′ UTR, respectively. Among these, 23 SNPs had been previous reported in public SNP databases (NCBI reference number, NT_006216.14). Six novel dCK SNPs were found (−2052C/A, IVS3 − 46G/del, IVS4 + 40G/T, IVS5 + 39T/C, IVS5 − 72A/T, and 966–975T10/T11) (Supplemental Table II). Two promoter SNPs of −360C/G and −201C/T have been reported previously in Caucasian, Japanese, and Chinese subjects. These two SNPs were in full linkage disequilibrium and found at an allele frequency of 26%. The allele frequencies of these two SNPs were more frequent than what has been reported in Caucasian, Japanese, and Chinese subjects (26% vs. 2%, 13.1% and 15.6%). Ten SNPs were found in introns (IVS1 + 37G/C, IVS1 − 174G/A, IVS1 − 110T/G, IVS2 + 114G/A, IVS3 − 46G/del, IVS3 − 45T/del, IVS4 + 40G/T, IVS5 + 39T/C, IVS5 − 72A/T, and IVS7 + 41A/T), of which IVS1 − 174G/A, IVS2 + 114G/A, and IVS7 + 41A/T were found at allele frequencies of 3%. Twelve SNPs were found in exons (A100A, P122S, 948T/C, 966–975T10/T11, 1796A/G, 1797A/G, 1827del/A, 1904del/C, 1952A/C, 2031A/G, 2105A/T, and 2254TT/del). One SNP was nonsynonymous and resulted in the substitution of proline at amino acid position 122 by serine (364C/T in exon 3, P122S). The allele frequencies of 948T/C and 966–975T10/T11 were 2%. The SNP located in the 3′ untranslated region of the gene was a synonymous SNP, and this SNP had an allele frequency of 2%. Linkage analysis showed significant disequilibrium between −360C/G and −201C/T (rho-square 1.0, P value <10−5).
DCK SNP Genotypes and Lamivudine Responses
Three dCK SNP genotype patterns (−360CC/−201CC, −360CG/−201CT, and −360GG/−201TT) were found among 127 CHB patients. Allele frequency analysis of −201C/T and −360C/G in 127 CHB patients as well as peripheral blood samples from 24 healthy volunteers and 24 patients with CHB revealed that these alleles were in Hardy–Weinberg equilibrium (HW P = 0.561). Sequence analysis exhibited that −360C/−201C and −360G/−201T were in complete linkage disequilibrium.
Patients did not show any differences in the distributions of dCK genotypes according to age (P = 0.551), and sex (P = 0.201). We next analyzed the relationship between dCK SNP genotypes and treatment responses. The 127 CHB patients were divided into two groups according to their responses to LAM monotherapy. Forty-seven patients achieved a good drug response, namely a virological response with undetectable HBV DNA for at least 12 months. Eighty patients showed a poor drug response, including 21 patients who did not have a decline in serum HBV DNA by ≥1 log10 IU/ml after the first 6 months of therapy and 59 patients whose serum HBV DNA increased by >1 log10 IU/ml (10-fold) above nadir after achieving virological response at 12 months. No obvious differences in clinical data such as sex, age, baseline serum ALT level, or HBV DNA level were found (Table I). There was no statistically significant difference between the two groups in terms of the distributions of SNP genotypes or allele frequencies (Table II). Multivariate analysis was performed using one categorized value (dCK SNP genotype) and three continuous values that were previously reported to be predictive of a good drug response. Age and baseline serum HBV DNA level were independent predictive factors of a good drug response (Table III).
| Variables | Good responsea (N = 47) | Poor responseb (N = 80) | P value |
|---|---|---|---|
| Male:Female (%) | 34:13 (72.3:27:7) | 64:16 (80.0:20.0) | 0.321 |
| Mean age (years) | 40.5 ± 11.1 | 46.0 ± 10.5 | 0.777 |
| ≤40 | 20 (42.6) | 21 (26.3) | 0.058 |
| >40 | 27 (57.4) | 59 (73.8) | |
| Baseline serum ALT (IU/L) | 224.8 ± 155.8 | 214.7 ± 189.2 | 0.431 |
| >2 and ≤5 times the upper limit of normalc | 24 (51.1) | 52 (65.0) | 0.122 |
| >5 times the upper limit of normalc | 23 (48.9) | 28 (35.0) | |
| Baseline HBV DNA (log10 copies/ml)d | 7.7 ± 1.0 | 8.0 ± 1.1 | 0.702 |
| ≤8.0 | 24 (51.1) | 31 (38.8) | 0.176 |
| >8.0 | 23 (48.9) | 49 (61.2) | |
| HBeAg-positive | 40 (85.1) | 74 (92.5) | 0.184 |
| Previous IFN-α therapy | 7 (16.7) | 5 (7.6) | 0.209 |
| Liver cirrhosis | 2 (4.3) | 8 (10.0) | 0.324 |
| Hepatocellular carcinoma | 0 (0) | 1 (1.3) | – |
| Mean treatment duration (months) | 30.0 ± 19.1 | 34.1 ± 18.8 | 0.240 |
| Mean follow-up duration (months) | 51.5 ± 20.5 | 47.3 ± 20.1 | 0.266 |
- Values are given as mean ± standard error of the mean. ALT, alanine aminotransferase; HBV, hepatitis B virus; IFN-α, interferon alpha.
- a Good response: virological response with undetectable HBV DNA for at least 12 months.
- b Poor response: virological response without a decline in serum HBV DNA ≥ 1 log10 IU/ml after the first 6 months of therapy and an increase in serum HBV DNA by >1 log10 IU/ml (10-fold) above nadir after achieving virologic response during continued treatment.
- c The upper limit of normal ALT levels is less than 40 IU/L.
- d The lower limit of detection for the HBV DNA test is 300 copies/ml.
| Genotype distribution | Allele frequency (%) | |||||
|---|---|---|---|---|---|---|
| Drug response | CC | CG + GG | P value | C | G | P value |
| Goodb (N = 47) | 32 (68.1) | 14 + 1 (31.9) | 0.243 | 78 (83.0) | 16 (17.0) | 0.251 |
| Poorc (N = 80) | 62 (77.5) | 17 + 1 (22.5) | 141 (88.1) | 19 (11.9) | ||
- a 127 patients with CHB who were treated with lamivudine were analyzed retrospectively.
- b Good response: virological response with undetectable HBV DNA for at least 12 months.
- c Poor response: virological response without a decline in serum HBV DNA ≥ 1 log10 IU/ml after the first 6 months of therapy and an increase in serum HBV DNA by >1 log10 IU/ml (10-fold) above nadir after achieving virologic response during continued treatment.
| Variables | Odds ratio | 95%CI | P value |
|---|---|---|---|
| G allele of −360C/G and T allele of −201C/T | 1.659 | 0.704–3.910 | 0.247 |
| Age (years) | 0.949 | 0.914–0.985 | 0.006 |
| Baseline serum ALT (IU/L)b | 1.000 | 0.998–1.002 | 0.909 |
| Baseline HBV DNA (log10 copies/ml)c | 0.682 | 0.468–0.994 | 0.047 |
- ALT, alanine aminotransferase; HBV, hepatitis B virus.
- a Good response: virological response with undetectable HBV DNA for at least 12 months.
- b The upper limit of normal ALT levels is less than 40 IU/L.
- c The lower limit of detection for the HBV DNA test is 300 copies/ml.
DCK SNP Genotypes and HBeAg Seroclearance
Among 127 patients, 13 patients with HBeAg-negative CHB were excluded. Three dCK SNP genotype patterns were found among the 114 HBeAg-positive CHB patients. Allele frequency analysis of −201C/T and −360C/G in the 114 HBeAg-positive CHB patients, as well as in peripheral blood samples from 24 healthy volunteers and 24 patients with CHB, revealed that these alleles were in Hardy–Weinberg equilibrium (P = 0.605).
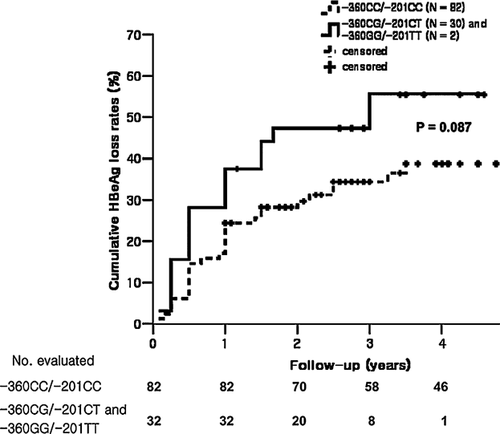
The 114 HBeAg-positive CHB patients were divided into two groups according to HBeAg seroclearance during LAM monotherapy. Forty-six patients achieved HBeAg seroclearance during the treatment period, whereas another 68 patients did not achieve HBeAg seroclearance. There were no obvious differences in sex, age, baseline serum ALT, or HBV DNA level among the two groups (Supplemental Table III). Interestingly, there was significant difference in the allele frequency of −360G/−201T between HBeAg seroclearance group and HBeAg non-seroclearance group (P = 0.045). (Table IV). However, multivariate analysis revealed that age was the only independent predictive factor for HBeAg seroclearance (Table V). Follow-up data concerning cumulative HBeAg seroclearance rates were available for 114 HBeAg-positive CHB patients. With a median follow-up of 51 (24–103) months, the 3-year cumulative HBeAg seroclearance rate for the −360CG/−201CT (N = 30) and −360GG/−201TT (N = 2) groups was 55.5%, whereas that for the −360CC/−201CC (N = 82) group was 31.2%. Cumulative HBeAg seroclearance rates of the two groups are presented in Figure 1. Although there were slightly more genotypes containing the −360G/−201T haplotype in the seroclearance group than the non-seroclearance group, there was no significant difference between the two groups (P = 0.087).
| Genotype distribution | Allele frequency (%) | |||||
|---|---|---|---|---|---|---|
| HBeAg status HBeAg seroclearanceb | CC | CG + GG | P value | C | G | P value |
| Yes (N = 46) | 29 (63.0) | 15 + 2 (37.0) | 0.082 | 73 (79.3) | 19 (20.7) | 0.045 |
| No (N = 68) | 53 (77.9) | 15 + 0 (22.1) | 121 (89.0) | 15 (11.0) | ||
- a 114 patients with HBeAg-positive CHB who were treated with lamivudine were analyzed retrospectively.
- b HBeAg clearance: achieving HBeAg seroclearance during continued treatment.
| Variables | Odds ratio | 95%CI | P value |
|---|---|---|---|
| G allele of −360C/G and T allele of −201C/T | 2.203 | 0.924–5.251 | 0.075 |
| Age (years) | 0.957 | 0.920–0.996 | 0.031 |
| Baseline serum ALT (IU/L)a | 1.001 | 0.999–1.003 | 0.396 |
| Baseline HBV DNA (log10 copies/ml)b | 0.695 | 0.457–1.056 | 0.088 |
| Mean treatment duration (months) | 1.013 | 0.991–1.035 | 0.246 |
- ALT, alanine aminotransferase; HBV, hepatitis B virus.
- a The upper limit of normal ALT levels is less than 40 IU/L.
- b The lower limit of detection for the HBV DNA test is 300 copies/ml.
DISCUSSION
Six novel dCK SNPs were found (−2052C/A, IVS3 − 46G/del, IVS4 + 40G/T, IVS5 + 39T/C, IVS5 − 72A/T, and 966–975T10/T11). Two promoters SNPs (−360C/G and −201C/T) that were previously reported in Caucasians, Japanese, and Chinese patients were found to be more frequent in our Korean patients [Shi et al., 2004; Joerger et al., 2006; Kim et al., 2008]. In addition, we found a SNP (P122S) in exon three that changes the amino acid sequence of the protein. This SNP in exon three has been found in healthy Caucasian volunteers, but not in Chinese cohort [Shi et al., 2004; Joerger et al., 2006; Lamba et al., 2007]. These findings show significant ethnic differences in SNPs of dCK gene. In previous reports, these genetic variations in dCK gene showed different gene expression and pharmacodynamic activities [Lamba et al., 2007]. In patients with acute myelogenous leukemia, Shi et al. reported that the patients with the −360CG/−201CT or −360GG/−201TT genotype expressed higher levels of dCK mRNA and showed a better response to chemotherapy than patients with the −360CC/−201CC genotype [Shi et al., 2004].
The change to pharmacologically active LAM metabolites requires sequential phosphorylation. Essentially, phosphorylation of LAM to LAM-monophosphate is catalyzed by deoxycytidine kinase. Deficiency in the activity of this enzyme might be associated with poor response to LAM as anticancer chemotherapeutic agents showed unfavorable response, whereas increased enzyme activity might be associated with increased activation and good response to LAM triphosphate derivatives. The relationships of pharmacogenetic factors to drug sensitivity and resistance are clinically important [Jacobsson et al., 1995; Antonelli et al., 1996; Groschel et al., 1997]. On the basis of previous reports, hypothesis was that CHB patients carrying the −360CG/−201CT or −360GG/−201TT genotype might achieve higher intracellular concentrations of LAM triphosphate and show a favorable response to drugs.
Unfortunately, when CHB patients were divided into two groups according to clinical response to LAM monotherapy, there was no significant difference between the two groups in terms of the distribution of SNP genotypes or allele frequencies. Although six novel dCK SNPs were discovered, these novel SNPs did not show any association with treatment response. However, in terms of HBeAg seroclearance, individuals with genotypes containing the −360G/−201T haplotype tended to show HBeAg seroclearance during the treatment period among patients with HBeAg-positive CHB. Cumulative HBeAg seroclearance rates showed a tendency to increase during long-term follow-up in CHB patients carrying the −360CG/−201CT or −360GG/−201TT genotypes. Especially, there was significant difference in the allele frequency of −360G/−201T between HBeAg seroclearance group and HBeAg non-seroclearance group (P = 0.045). The possible explanation of our results is that the patients with the −360G/−201T haplotype may exhibit a series of sequential effects: First, the patients may have higher levels of dCK mRNA. Second, higher levels of dCK mRNA may induce effective LAM metabolism, and lastly, the effective LAM metabolites may contribute to HBeAg seroclearance.
The main concern with long-term NA therapy is the emergence of drug resistance. Long-term treatment of CHB patients with NA agents may result in failure of therapy because of the emergence of virus variants with decreased susceptibility to NA agents. The change of drug metabolism in host cells may contribute to inefficient LAM phosphorylation in CHB patients. Thus, intracellular sub-therapeutic levels of the active compounds may develop. Altered intracellular environment may induce the selection of virus variants with resistance [Zoulim, 2004; Shaw et al., 2006]. Host factors as well as viral resistance may attenuate the efficacy of antiviral agents.
Although the current first line NAs, entecavir and tenofovir, have high potency and high barriers to resistance, HBV cannot be completely eradicated. Most guidelines recommend continuous antiviral therapy to patients with CHB until HBsAg seroclearance. Therefore, there is an unmet need for genotyping of disease specific SNPs to determine prognosis and to lead optimizing personalized therapy [European Association For The Study Of The L, 2012]. Pharmacogenomics would play an important role in preventing patients from the development of drug resistance. Based on our results, enzymatic activation of LAM probably has an effect on antiviral activity in HBeAg-positive CHB patients in terms of HBeAg seroclearance. Nevertheless, in multivariate analysis, the G allele of −360C/G and the T allele of −201C/T were not significant predictive factors for HBeAg seroclearance or a good response. These findings suggest that other host and viral factors, including age, viral load, and YMDD mutations have much more influence on the efficacy of treatments for CHB.
A limitation of this study is the small sample size. Preferentially the validation in another cohort receiving LAM or in other cohort receiving other NAs would be helpful. If correlations could be confirmed in a larger number of patients, the SNPs we identified in the dCK gene could potentially be used as markers to predict treatment response to antiviral agents. Second, the functional studies of dCK candidate SNPs should be conducted in CHB patients receiving antiviral agents. They would clarify whether these candidate SNPs are associated with the expression of dCK mRNA, as well as the level of active NAs triphosphates. Lastly, the quantification of anti-HBe or HBsAg level should have clarified that −360CG/−201CT or −360GG/−201TT genetic variation might help HBeAg or HBsAg seroclearance.
In conclusion, we found 29 dCK SNPs. Among these, six dCK SNPs were novel. Two promoter SNPs, namely −360C/G and −201C/T, had a higher frequency in Koreans than other ethnic groups. Although neither the genotype nor the allele frequency of −360G/−201T were significant predictors of treatment response, there was significant difference in the allele frequency of −360G/−201T between HBeAg seroclearance group and HBeAg non-seroclearance group (P = 0.045). These results suggest that HBeAg-positive CHB patients with the −360G/−201T haplotype might help HBeAg seroclearance in response to LAM treatment.
A better understanding regarding the functional polymorphisms of candidate gene provides significant directions to discover the reason of primary non-response and treatment failure unrelated to viral mutations during highly potent antiviral therapy. Pharmacogenomics may help physicians to accurately recommend effective antiviral agents to individual patients. Although LAM is no longer a first-line antiviral agent, the concepts proposed here are applicable to other antiviral agents. Our data provide important insights into the relationships between dCK gene polymorphisms and the efficacy of antiviral agents.
ACKNOWLEDGMENTS
Hyun Woong Lee analyzed the clinical data, performed research, and wrote the paper; Sung Hee Lee, Hye Young Chang, and Min Goo Lee performed research; Sang Hoon Ahn analyzed the clinical data and coordinated the research project; Kwang-Hyub Han was instrumental in developing and coordinating the research project and reviewed the manuscript.




