Surveillance of Adenovirus D in patients with epidemic keratoconjunctivitis from Fukui Prefecture, Japan, 1995–2010
Abstract
Human adenoviruses species D (HAdV-D) are known to cause severe epidemic keratoconjunctivitis. However, the isolation rate of HAdV-D is not high, because HAdV-D is usually slow to propagate. Although new types of HAdV-D have been reported, accurate surveillance has not been performed because of difficulties in culturing the viruses and lack of a practical identification method. In this study, HAdV-Ds were detected and identified from patients with epidemic keratoconjunctivitis in the Fukui Prefecture during 1995–2010 by PCR, loop-mediated isothermal amplification (LAMP) of DNA, and conventional virus isolation and neutralization tests. All samples were subjected to culture and PCR and LAMP. A total of 124 strains of HAdV-D were detected from 157 patients with epidemic keratoconjunctivitis. The strains consisted of the following types: D8 (n = 8), D19 (n = 4), D37 (n = 40), D53 (n = 5), D54 (n = 66), and D56 (n = 1). Among these, D53, D54, and D56 are new types that have been reported recently. The results of this study demonstrated that new types of HAdV-D caused epidemic keratoconjunctivitis during 1995–2010, and included an outbreak of keratoconjunctivitis caused by HAdV-D54. The LAMP method was able to detect and identify HAdV-D53 and HAdV–D54 in 1 hr, and may therefore be applicable for use at the bedside. J. Med. Virol. 84:81–86, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Human adenoviruses (HAdVs) cause a wide range of infections, including respiratory infections, eye diseases, and gastroenteritis [Wold and Marshall, 2007]. HAdVs are divided into seven species, A to G [Jones et al., 2007]. Among these species, human species D adenovirus (HAdV-D) includes several types that cause epidemic keratoconjunctivitis. HAdV-D type D8, -D19a, and -D37 are well known to cause severe epidemic keratoconjunctivitis, whereas the other types of HAdV-D usually do not [Aoki and Tagawa, 2002].
Recently, new types of HAdV-D have emerged as causative agents of severe epidemic keratoconjunctivitis. These include HAdV-D53 [Aoki et al., 2008; Walsh et al., 2009; Kaneko et al., 2011a], -D54 [Ishiko et al., 2008; Ishiko and Aoki, 2009; Kaneko et al., 2011b], and -D56 [Kaneko et al., 2011c; Robinson et al., 2011]. Like HAdV-D8, some types of HAdV-D are also fastidious and difficult to isolate from clinical samples [Kaneko et al., 2011b]. Moreover, HAdV-D types have cross-neutralizing reactions with other HAdV-D [Hierholzer et al., 1991]. Therefore, in this study, gene-based methods such as polymerase chain reaction (PCR) [Miura-Ochiai et al., 2007; Robinson et al., 2011] and loop-mediated isothermal amplification (LAMP) [Notomi et al., 2000; Wakabayashi et al., 2004] were used to identify HAdV-Ds.
MATERIALS AND METHODS
Patients and Clinical Samples
A total of 124 strains of HAdV-D were detected in 157 patients with epidemic keratoconjunctivitis over a 16-year period from 1995 to 2010. Eye scrapings were collected in a virus surveillance system established and approved in Fukui prefecture (population, 821,589 in 2005) Japan. Conjunctival swabs were transferred into a test tube containing 1 ml of Eagle's. MEM containing 0.5% bovine serum albumin were used as a clinical sample for a later laboratory diagnosis. The test tubes were kept at −80°C until use. The number of patients by year are as follows: 1995 (n = 5), 1996 (n = 5), 1997 (n = 5), 1998 (n = 1), 1999 (n = 0), 2000 (n = 8), 2001 (n = 7), 2002 (n = 7), 2003 (n = 8), 2004 (n = 17), 2005 (n = 51), 2006 (n = 19), 2007 (n = 10), 2008 (n = 0), 2009 (n = 4), and 2010 (n = 10). In 2004, 13 out of 17 patients with epidemic keratoconjunctivitis were suspected cases of nosocomial infection. In this report, HAdV-D indicates family Adenoviridae, genus Mastadenovirus, and species Human adenovirus D. All samples were subjected to culture and PCR and LAMP.
Viral Nucleic Acid Extraction
QIAamp MinElute Virus Spin Kits (Qiagen, Hilden, Germany) or QIAamp Viral RNA Mini Kits were used to extract viral nucleic acid from clinical samples. The former used 200 µl of clinical sample and the latter used 140 µl. Viral nucleic acid was eluted in 60 µl of elution buffer. A MinElute Virus Spin Kit was used if there was a sufficient sample volume.
Viral Culture and Neutralization
Clinical samples were inoculated onto MRC-5, Caco-2, HEp-2, and Vero-E6 cells as reported previously [Enomoto et al., 2010]. After inoculation, these cells were subpassaged three times, once a week. Neutralization tests were performed using antisera against HAdV-D8 (ATCC, Rockville, MD), HAdV-D19 (Denka, Tokyo, Japan), and HAdV-D37 (Denka).
PCR Sequencing
PCR and sequencing were performed using the primers AdnU-S′2 and AdnU-A2 [Miura-Ochiai et al., 2007]. The primer sequences were as follows: AdnU-S′ 2 [5′-TTC CCC ATG GCN CAC AAY AC-3′, N (A/T/C/G), Y (C/T)] and AdnU-A2 [5′-TGC CKR CTC ATR GGC TGR AAG TT-3′, K (G/T), R (A/G)]. An aliquot (2 µl) of each extracted viral DNA sample was used as the template. Amplification reactions were conducted in 50 µl of reaction mixture containing 0.5 µM each of primers, 200 µM each of dideoxynucleotides, 1 U Speed Star HS DNA polymerase (TaKaRa Shuzo, Shiga, Japan), 10 mM Tris–HCl (pH 8.0), and 50 mM KCl. PCR amplification (GeneAmp PCR System 9700, Applied Biosystems, Foster City, CA) consisted of 40 cycles of 95°C for 5 sec, 50°C for 15 sec, and 72°C for 10 sec. PCR products (554 bp) were purified using QIAquick PCR purification kits (Qiagen) and used as templates for DNA sequencing reactions.
Partial hexon gene sequences (350 bp) were determined and used to type the HAdVs. The locations of the primers and target regions on HAdV-D genes are shown in Figure 1. To sequence other HAdV-D regions, PCR sequencing primers were designed with reference to the complete genome sequences of other HAdV-Ds registered previously in the GenBank/EMBL/DDBJ databases.
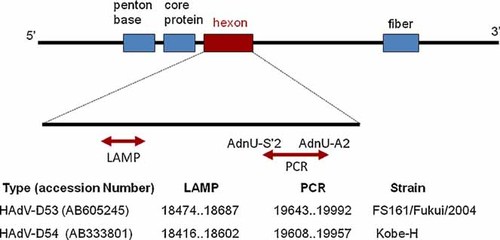
Locations of the primers and target regions on human adenovirus D genomes [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com].
Nucleotide sequencing was performed using an autosequencer (Applied Biosystems 3130 Genetic Analyzer, Foster City, CA) and a commercial kit (BigDye Terminator V3.1 Cycle Sequencing Kit; Applied Biosystems). The nucleotide sequences on both strands of the amplicons were determined by sequencing the target regions.
LAMP
Two sets of type-specific primers were designed for HAdV-D53 and HAdV-D54 (Table I). The primers used for HAdV-D53 were F3-02, B3-01, FIP-02, BIP-03, and LB-03, and those for HAdV-D54 were F3-02′, B3-02, FIP-02-2, BIP-02, LF-01, and LB-02. LAMP [Notomi et al., 2000] was carried out using Loopamp DNA Amplification Kits (Eiken Chemical Co., Ltd, Tokyo, Japan). Each 25 µl reaction mixture contained 2 µl of extracted DNA, 12.5 µl of 2× reaction mixture, 8 units of Bst DNA polymerase, 0.4 µM each of the primers F3 and B3, 1.6 µM each of FIP and BIP, and 0.8 µM each of LF and LB; and was incubated at 65°C for 60 min and then heated at 80°C for 2 min. Real-time turbidity was monitored using an LA-320C turbid meter (Eiken Chemical Co., Ltd) or similar LAMP instrument. LAMP Target regions are shown in Figure 1.
| Target type | Primer type for LAMP | Primer name | Sequence (5′ → 3′) | Reference strain | Position based on reference strain | Polarity |
|---|---|---|---|---|---|---|
| HAdV-D53 | F3 | F3-02 | AACTAATGCCGAAGGTCA | Human adenovirus 53 DNA, complete genome. Isolate FS161/Fukui/2004, 35116 bp, accession no. AB605245 | 18474–18491 | Positive |
| B3 | B3-01 | GTTGGGCATGGACTGCTG | 18687–18670 | Negative | ||
| FIP | FIP-02 | GGTTAAATTCATTATCACCTGCCAGAGGAGTTAGACATTGACC | 18565–18543 | Negative–positive | ||
| 18498–18517 | ||||||
| BIP | BIP-03 | ATATGAATCTGGAGACGCCAGACGCTAAGTTAGCTTCAGAAC | 18590–18611 | Positive–negative | ||
| 18669–18650 | ||||||
| LB | LB-03 | CAAACCTGGAACTTCAGATGACA | 18627–18649 | Positive | ||
| HAdV-D54 | F3 | F3-02′ | AAAACTGGAAATGATGGCC | Human adenovirus 54 DNA, complete genome. Isolate Kobe-H, 34920 bp, accession no. AB333801 | 18416–18434 | Positive |
| B3 | B3-02 | AACTGTCTTCTAAAGGTCCT | 18602–18583 | Negative | ||
| FIP | FIP-02-2 | TGTCTTCAGCATTAGTGTCGCAGCATGACATAACAATGGC | 18503–18483 | Negative–positive | ||
| 18447–18465 | ||||||
| BIP | BIP-02 | GAAGCAGACATTGTTATGTACACCGGCTTGTACACCACATGA | 18512–18535 | Positive–negative | ||
| 18582–18565 | ||||||
| LF | LF-01 | CAGGAGTATCAAAGAAA | 18482–18466 | Negative | ||
| LB | LB-02 | GTTAATCTTGAAACTCCAGATAC | 18542–18564 | Positive |
Experimental Ethics
Clinical samples were collected and evaluated during routine pathogen surveillance, which is performed in Japan under “The Law Concerning the Prevention of Infectious Diseases and Medical Care for Patients of Infections (the Infectious Diseases Control Law)”. Informed consent was obtained from all patients.
RESULTS
Virus Isolation and Identification
HAdVs were isolated and identified as HAdV-D19 (n = 3), HAdV-D37 (n = 36) by culture and neutralization test. HAdV-D53 (n = 4), and HAdV-D56 (n = 1) were isolated and identified by PCR and sequencing and/or LAMP methods. No HAdV-D8 or HAdV-D54 types could be isolated.
HAdV-D53 and HAdV-D56 have been reported as HAdV-22/37/H8 [Aoki et al., 2008] and HAdV-15/29/H9 [Kaneko et al., 2011c], respectively. In this report we used the terms HAdV-D53 and HAdV-D56 because these names are accepted on a worldwide basis [Aoki et al., 2011; Seto et al., 2011].
Results of PCR and Sequencing
A total of 121 HAdV-D strains were detected by PCR sequencing. HAdV-D strains were typed as HAdV-D8 (n = 8), HAdV-D19 (n = 4), HAdV-D37 (or-D53) (n = 45), HAdV-D54 (n = 63), and HAdV-D56 (n = 1) (Table II).
| LAMP method | Total | |||
|---|---|---|---|---|
| HAdV53/22 positive | HAdV54 positive | Both negative | ||
| PCR-sequencing | ||||
| HAdV-D8 | 8 | 8 | ||
| HAdV-D19 | 4 | 4 | ||
| HAdV-D37/53 | 5 | 40 | 45 | |
| HAdV-D54 | 63 | 63 | ||
| HAdV-D56 | 1 | 1 | ||
| Negative | 3 | 3 | ||
| Total | 5 | 66 | 53 | 124 |
Complete genome sequences of HAdV-D53 strains were determined [Kaneko et al., 2011a] and deposited in GenBank with accession numbers AB605245 and AB605246. Partial sequences of HAdV-D56 were determined and deposited in GenBank with accession numbers AB630324 and AB630325. The deposited HAdV-D56 sequences had 100% identity to previously deposited sequences of HAdV-D56 (HM770721) [Robinson et al., 2011].
LAMP for HAdV-D53 and HAdV-D54
Among the HAdV-D37 (or D-53) (n = 45) samples, 5 were identified as HAdV-53 by LAMP. All the isolates (n = 36) identified as HAdV-D37 by neutralization testing were negative by LAMP in this study. By using LAMP specific for HAdV-D54, HAdV-D54 genomes were identified in 66 patients. All 63 samples identified as HAdV-D54 by PCR sequencing [Miura-Ochiai et al., 2007] were also positive by LAMP, which identified 3 additional samples as HAdV-D54 strains (Table II).
HAdV-D Types Isolated by Years
HAdV-D53 was first isolated in 1996, and there have been no large outbreaks associated with this type. HAdV-D54 and HAdV-D56 first appeared in 2000 and 2010, respectively, in Fukui prefecture. Among these new types, HAdV-D54 caused large outbreaks of epidemic keratoconjunctivitis from 2005 to 2006. HAdV-D37 has been isolated continuously and has caused epidemic keratoconjunctivitis. In 2004, 2007, and 2010, HAdV-D37 was the main causative agent of epidemic keratoconjunctivitis. HAdV-D8 has not been isolated since 2002, and HAdV-D19 has not been isolated since 2006 (Table III).
| Year | Types | Total | |||||
|---|---|---|---|---|---|---|---|
| HAdV-D8 | HAdV-D19 | HAdV-D37 | HAdV-D53 | HAdV-D54 | HAdV-D56 | ||
| 1995 | 1 | 1 | |||||
| 1996 | 1 | 2 | 1 | 4 | |||
| 1997 | 3 | 1 | 4 | ||||
| 1998 | 0 | ||||||
| 1999 | 0 | ||||||
| 2000 | 1 | 1 | 2 | ||||
| 2001 | 4 | 1 | 5 | ||||
| 2002 | 0 | ||||||
| 2003 | 1 | 2 | 1 | 4 | |||
| 2004 | 9 | 3 | 2 | 14 | |||
| 2005 | 1 | 2 | 45 | 48 | |||
| 2006 | 3 | 15 | 18 | ||||
| 2007 | 9 | 1 | 10 | ||||
| 2008 | 0 | ||||||
| 2009 | 3 | 1 | 4 | ||||
| 2010 | 9 | 1 | 10 | ||||
| Total | 8 | 4 | 40 | 5 | 66 | 1 | 124 |
DISCUSSION
HAdV-D is the most important causative agent of epidemic keratoconjunctivitis [Aoki and Tagawa, 2002; Wold and Marshall, 2007]. A total of 124 strains of HAdV-D consisting of six types were detected from patients with epidemic keratoconjunctivitis in Fukui Prefecture from 1995 to 2010. Conventionally, typing of HAdV is done by neutralization tests [Wold and Marshall, 2007]. Neutralization of a virus is defined as the loss of infectivity through reaction of the virus with specific antibody; however, cross-reactivity among HAdV-D serotypes has been reported [Hierholzer et al., 1991]. Since 2008, the term “type” began to be used internationally instead of “serotype.” The new HAdVs have been determined by complete genome sequences [Aoki et al., 2011; Seto et al., 2011], and include HAdV-D53, HAdV-D54, and HAdV-D56, which were identified and reported based on the sequencing of their entire genomes. All these have caused epidemic keratoconjunctivitis in Japan.
HAdV-D53 was classified as serotype 22 by neutralization testing. However because of epitope sharing, neutralization testing could not differentiate HAdV-D53 from the prototype strain of HAdV-D22. HAdV-D53 causes epidemic keratoconjunctivitis, whereas the prototype of HAdV-D22 does not [Walsh et al., 2009]. HAdV-D22 strains isolated from patients with epidemic keratoconjunctivitis in Japan were found to be intertypic recombinant strains of HAdV-D22 by genetic analysis [Kaneko et al., 2011a] and were designated HAdV-D53. Although LAMP designed for HAdV-D53 amplifies the hexon gene of HAdV-D22, since the HAdV-D22 does not cause epidemic keratoconjunctivitis [Walsh et al., 2009], if LAMP for HAdV-D53 is positive in patients with epidemic keratoconjunctivitis, HAdV-D53 infection should be considered.
HAdV-D54 is both a new type and new serotype. Before 2008, HAdV-D54 was misidentified as a variant strain of HAdV-D8 [Ishiko et al., 2008; Ishiko and Aoki, 2009; Kaneko et al., 2011b], because of cross neutralization with HAdV-D8. In Fukui prefecture, the same viral culture and neutralization methods have been used for identification of HAdV-Ds. Since 2004, the isolation rate of HAdV-Ds from epidemic keratoconjunctivitis cases began to decrease because the causative agent was HAdV-D54, which is too fastidious to isolate by culture. From 2004 to 2006, a total of 62 strains of HAdV-D54 were detected by PCR and/or LAMP, and LAMP was therefore thought to be a sensitive, rapid and practical method for identifying both HAdV-D53 and HAdV-D54. LAMP was also found to be as sensitive as nested-PCR sequencing [Miura-Ochiai et al., 2007]. PCR sequencing can detect all types of HAdVs, but it is usually time consuming. LAMP [Notomi et al., 2000] is potentially applicable as a bedside diagnostic method, because it does not require specific instruments and rapidly provides results. Very rapid PCR methods [Fujimoto et al., 2010] have also been developed, but at present, these cannot identify all HAdV types, because the PCR products are too short.
Kaneko et al. [2011b] reported that they had not detected any HAdV-D8 strains since 2004 in Japan, and the same trend has also been observed in Fukui prefecture. There was no HAdV-D8 detected from 2002 to 2010. HAdV-D53 was first isolated in 1996 in this study, but no outbreak has been observed. HAdV-D56 [Robinson et al., 2011] was isolated in 2010, and the sequence was identical to the novel intertypic recombinant, HAdV-15/29/H9 [Kaneko et al., 2011c].
Six different types of HAdV-D were the causative agents of epidemic keratoconjunctivitis in Fukui prefecture during 1995–2010. During 2005–2006, HAdV-D54 was identified from an outbreak of epidemic keratoconjunctivitis by PCR and/or LAMP methods. HAdV-D54 causes severe epidemic keratoconjunctivitis and induces corneal opacities that can persist for over 1 year [Akiyoshi et al., 2011], leading to decreased quality of life. Early diagnosis of HAdV-D54 is thought to be important for outbreak prevention and patient management.
This study demonstrates that, in addition to conventional serological testing, supplemental diagnostic methods such as PCR and LAMP applications enable better HAdV-D surveillance. LAMP applications in this study used five primers for HAdV-D53 and six primers for HAdV-D54. These primers recognize seven and eight regions, respectively. LAMP requires many specific genomic sequences and holds higher specificity than PCR, which utilizes only two genomic regions. In addition, a subsequent sequencing procedure is not needed with this method. Therefore, LAMP is more rapid than PCR sequencing. Additional studies using LAMP should be considered for other types of HAdV.
Acknowledgements
We thank public health officers of health centers in Fukui prefecture and hospital staffs for sample collection.




