Transcriptional features and transcript structure of UL145 in different strains of human cytomegalovirus
Abstract
The human cytomegalovirus UL145 gene is located between the highly variable genes UL144 and UL146 in the UL/b′ region. However, unlike its neighboring genes, UL145 is relatively conserved among strains isolated from patients. The transcriptional features and transcript structure of UL145 has not yet been examined. In this study, the transcriptional features and structure of UL145 were characterized using RNA preparations from three HCMV strains isolated from patients, designated H, C, and X, respectively. Two transcripts were identified by cDNA library screening. Two main clusters of transcripts, one located between 500 and 700 nt, and the other between 1,400 and 1,700 nt, were confirmed by Northern blot analysis. Abundant transcripts were detected at 96 h post-infection by Northern blot, suggesting that UL145 was a late gene. The terminal sequences of transcripts obtained by 3′ rapid amplification of cDNA ends demonstrated the presence of polyadenylation. Transcripts identified in the RNA of H, C, and X strains 96 h post-infection had the same 3′ end but different 5′ ends. In addition to polycistronic transcripts with upstream genes, UL145 could be transcribed as a single transcript during late strain infections. J. Med. Virol. 83:2151–2156, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Human cytomegalovirus (HCMV) is the prototypic β-herpesvirus and may cause severe disease both in neonates and immunocompromised individuals, such as allograft recipients and AIDS patients [Jenkins et al., 2008]. As one of the largest known human viruses, the HCMV genome comprises 236 kb of double-stranded DNA, and many of its genes are non-essential for replication in fibroblasts [Davison et al., 2003; Dunn et al., 2003]. The UL145 gene is in the 15 kb unique region of the genome termed UL/b′, which contains at least 19 open reading frames (ORFs). The UL/b′ region is found in HCMV isolates, but not in laboratory strains passaged extensively and is not necessary for growth in vitro [Chee et al., 1990; Cha et al., 1996]. Recent evidence demonstrates that clinical isolates, but not the attenuated laboratory strain AD169, can replicate actively in SCID mice which have been recipients of human xenografts, suggesting that the UL/b′ region is required for viral infection in vivo [Mocarski et al., 1993; DiLoreto et al., 1994; Wang et al., 2005].
The HCMV UL145 gene lies between the UL144 and UL146 genes, which have been demonstrated to be highly polymorphic among field isolates from different patients [Clark-Lewis et al., 1994; Benedict et al., 1999; Lurain et al., 1999; Penfold et al., 1999; Hassan-Walker et al., 2004]. Previous studies showed that the sequences from HCMV UL144 and UL146 were highly variable in clinical strains in China [He et al., 2004, 2006]. But unlike its neighboring genes, UL145 is relatively conserved in all strains, irrespective of the strain source. Data show that the putative protein kinase C (PKC) phosphorylation motif and casein kinase II (CK II) phosphorylation site are highly conserved in UL145 putative proteins of strains. The relative conservation of UL145 sequences being retained under selection pressure suggested that it might be essential for virus survival and replication in vivo [Sun et al., 2007]. Murphy et al. [2003] predicted that a total of 252 ORFs, including UL145, have the potential to encode proteins. However, there has been no report to date of the transcript structure of UL145 in HCMV strains isolated from patients. This study was therefore designed to characterize the transcriptional features and transcript structure of the UL145 gene in HCMV strains isolated from patients.
METHODS
Cells and Virus
Human embryo lung fibroblasts (HELFs) were used to culture HCMV for both production of viral stocks and HCMV RNA for later analysis. HELFs were cultured in 1640 medium supplemented with 15% fetal calf serum (FCS). Three strains, termed H, C, and X, were isolated from urine samples of neonates admitted to the Pediatrics Department, Affiliated Shengjing Hospital of China Medical University. These three patients were at ages of <5 months. The patient infected by H suffered from cytomegalovirus hepatitis, congenital biliary atresia and congenital heart disease. The patient infected by C lead to pertussis syndrome and thrush. The patient infected by X suffered from epilepsy and acute bronchopneumonia. HELFs were inoculated with the three viral strains at a multiplicity of infection (MOI) of 3–5 and the infected cells were collected for RNA preparation.
RNA Preparation
To obtain viral immediate-early (IE) RNA, cycloheximide (CHX) (100 µg/ml) was added to the culture medium 1 h prior to infection and the cells were harvested 24 h post-infection (hpi). For early (E) RNA preparation, phosphonoacetic acid (PAA) (100 µg/ml) was added immediately after infection and the cells were harvested at 48 hpi. Late (L) RNA was derived from infected cells and harvested at 96 hpi. Total RNA from fresh HCMV-infected cells was isolated with Trizol reagent (Invitrogen, Carlsbad, CA) and treated with DNA-free reagent (Ambion, Austin) to remove contaminating genomic DNA.
Screening of UL145 cDNA Clones in a cDNA Library
A full-length cDNA library of L RNA from HCMV strain H had been constructed previously in pBluescript II SK vector by the SMART technique (Clontech) [Ma et al., 2011]. The primary library consisted of 1.12 × 106 recombinant clones/ml. Original recombinants of the cDNA library were transferred into Escherichia coli DH5α cells. A total of 4,000 single clones were randomly picked and inoculated into LB medium. Aliquots of every 10 single clones were mixed together as the first grade of colonies, so were the second and third grade of colonies. Every thousand clones were classed as one group, named A, B, C, D, respectively. DNA of the propagated E. coli was exposed by using cell lysis buffer (50 mM TrisCl pH 6.8, 15 mM NaCl, 5 mM EDTA, 0.5% NP-40). To identify UL145 gene-specific clones, PCR was performed using each of the DNA preparations as templates sequentially from the third, second, first grades of colonies and the single clone. Gene-specific primers, 145U1 and 145D1 (shown in Table I) were used in the PCR screening. The PCR cycles were as follows: initial denaturation at 94°C for 5 min, 30 cycles of 94°C for 30 sec, 58°C for 45 sec, and 72°C for 1 min, followed by final elongation at 72°C for 10 min. Identified clones were sequenced using the T7 primer of the pBluescript II SK vector, using the ABI PRISM 3730 DNA analyzer (Applied Biosystems, Carlsbad, CA).
| Primers | Sequence (5′–3′) | Positionsa |
|---|---|---|
| cDNA library screening | ||
| 145U1 | CATCGCAAAGAAAGTCCC | 12,539 |
| 145D1b | GTCATCGTCGCTCCAAAC | 12,717 |
| Northern blotc | ||
| 145 norF | TCATCAGTAGTCCCAGCGTTAT | 12,383 |
| 145 norR | TAATACGACTCACTATAGGGTCTAGCTTGCGGAGCATCTC | 12,632 |
| 3′ RACE | ||
| 3′ RACE outer primer | GGCGTCCTGGCTCATTACTA | 12,358 |
| 3′ RACE inner primer | CGTCGCAAAGAAAGTCCCT | 12,539 |
| 5′ RACE | ||
| 5′ RACE inner primer | GGCTGTCACGGCACTGTAT | 12,588 |
- a Positions of 5′ termini of each primers corresponding to the H strain (GenBank accession No. GQ981646).
- b Not only used as reverse primer for cDNA library screening but also used as 5′ RACE outer primer.
- c Used as forward and reverse primer for amplification of the template for probe synthesis.
Identification of UL145 Transcripts by Northern Blot
RNA preparations of H, C, and X strains in IE, E, and L kinetics were used in Northern blot hybridization using the digoxigenin (DIG) Northern Starter Kit (Roche, Indianapolis, IN). Ten micrograms of RNA per lane were separated by electrophoresis on a 1.2% formaldehyde agarose gel and transferred to a positively charged nylon membrane. The quantities of 28S and 18S rRNAs in each RNA preparation were used as loading controls. The RNA probe for detection of UL145-specific RNA by Northern blot was generated by PCR amplification with the specific primers, 145 norF and 145 norR (Table I). The UL145-specific probe was labeled using DIG according to the manufacturer's protocol. Nylon membranes were prehybridized at 55°C in preheated hybridization solution supplied in the kit for 30 min. Labeled probe was first denatured by heating at 100°C for 5 min and then added to the prehybridized membrane and incubated overnight at 55°C. Post-hybridization, membranes were washed twice with 2× SSC, 0.1% SDS buffer, and 0.1× SSC, 0.1% SDS buffer at 55°C under constant agitation. After incubation with an anti-DIG antibody labeled with alkaline phosphatase, the blots were visualized by addition of the substrate of CDP-Star for 10 min.
Determination of UL145 cDNA Ends by Rapid Amplification of cDNA Ends (RACE)
In order to obtain the 5′ and 3′ ends of UL145 cDNA sequences, 5′ RACE and 3′ RACE were performed by using 5′-Full Race Kit and 3′-Full RACE Core Set Ver.2.0 kits (TaKaRa, Dalian, China) following the protocol supplied by the manufacturer. Briefly, the 3′ RACE PCR was carried out using L RNA preparations of H, C, and X strains. First-strand cDNA was synthesized with M-MLV reverse transcriptase and 3′ RACE Adaptor supplied in the kit, which contained oligo-dT and adaptor primer-binding sequence. Nested PCR was performed using the cDNA as template and the UL145-specific 3′ RACE outer and inner primers (Table I). Reactions were carried out as follows: 94°C for 3 min, 20 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 1 min, final elongation at 72°C for 10 min.
Similarly, 5′ RACE was carried out by using the L RNA preparations of H, C, and X strains. According to the protocol, RNA was treated with alkaline phosphatase to avoid non-specific ligation of incomplete mRNA, tRNA, rRNA, and contaminating genomic DNA with the adaptor. Tobacco acid pyrophosphatase was used to remove the 5′ cap structure from full-length mRNA, leaving a 5′-monophosphate. Then, the 5′ RACE adaptor was ligated to the mRNA with 5′-monophosphate using T4 RNA ligase. The mRNA, which contained the adaptor, was used as template for reverse transcription by M-MLV. After reverse transcription, nested PCR was performed using the gene-specific outer and inner primers (Table I). The cycles were as follows: 94°C for 3 min, 20 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 1 min, followed by final elongation at 72°C for 10 min. These gene-specific primers were designed based on the sequence data of the HCMV H strain (GQ981646). All RACE products were gel purified and cloned using the pCR 2.1 TA cloning kit (Invitrogen) and sequenced using M13F and M13R primers.
RESULTS
Identification of the UL145 Transcript by cDNA Library Screening
By screening a cDNA library by PCR and DNA sequencing, two cDNA clones were identified containing UL145-specific sequences. The lengths of the inserted sequences in the two clones were 1,459 and 608 bp, respectively. Compared with the sequence of the HCMV H strain (GQ981646), both ended with a polyA tail located downstream of the UL145 ORF. A polyA signal (AATAAA) was found in the cDNA sequences at 12,821 nt. The longer sequence stretched from nucleotide 11,400 to 12,843, with the 5′ end being located at −184 bp upstream of the UL144 ORF. The shorter sequence ranged from nucleotide 12,256 to 1,2845, with the 5′ end −99 bp upstream of the UL145 ORF. The sequences of the two cDNA clones have been submitted to the Genbank: ID HQ537475-HQ537476.
Identification of UL145 Transcript by Northern Blot
Two main clusters of transcripts were identified in all the three strains by Northern blot (Fig. 1). One was located between 500 and 700 nt, and the other between 1,400 and 1,600 nt. However, the kinetics and quantities of the transcripts were not completely consistent among the three strains. For strains H and C, the two clusters of transcripts were detected in both E and L RNA, but not in IE RNA, with the transcripts in L RNA being much more abundant than in E RNA. Alternatively, the transcripts were found only in L RNA for the X stain. In addition, the transcripts located between 500 and 700 nt were more dominant than those located between 1,400 and 1,600 nt in the L RNA of the H strain.
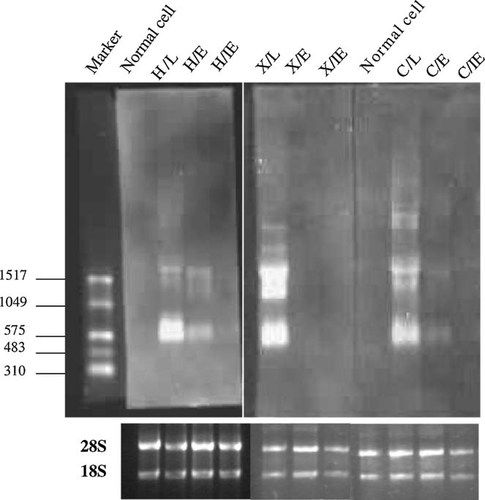
Northern blot analysis of UL145 transcripts. To equalize the amounts of RNA loading, the quantities of 28S and 18S rRNAs in each RNA preparations were determined by EB staining in a separate agarose gel.
Determination of 5′ and 3′ Ends of the UL145 Transcripts by Rapid Amplification of cDNA Ends
To identify precisely the 5′ and 3′ ends of the UL145 transcripts, RACE reactions were performed using L RNAs of the three strains. The results of 3′ RACE showed that the transcripts of UL145 in the H, C, and X strains had the same 3′ end at 12,848 nt, compared with HCMV H strain (Fig. 2). Consistent with the results of cDNA library screening, all the transcripts had the same polyA signal sequence (AATAAA) at 12,821 nt.
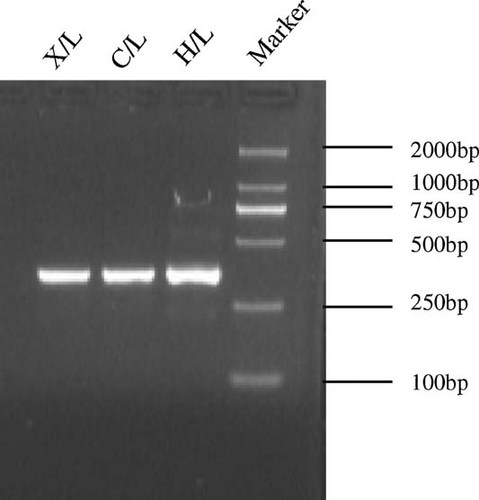
3′ RACE of UL145 of strains H, C, and X in L expression kinetics.
Several bands were obtained in L RNAs of the three strains by 5′ RACE (Fig. 3). They presented in two size ranges of 250–500 bp and 1,000–2,000 bp. All the products within 250–500 bp were gel purified, cloned and sequenced. Compared with the sequence of H strain (GQ981646), one 5′ end of cDNA sequence at 12,335 nt was identified in the L RNAs of H and X strains, and an adjacent 5′ end at 12,339 nt was found in the L RNA of the C strain. In addition, multiple 5′ ends were found in the three strains. They were distributed into two regions between 12,269 and 12,290 nt and from 12,210 to 12,191 nt, respectively. All of these 5′ends were located within the un-coding region between UL144 and UL145 ORFs. Partial products within 1,000–2,000 bp were sequenced and showed that they were unspliced sequences containing the sequences of UL144 and UL145 ORFs. Multiple 5′ ends were found including that of the 1,459 bp cDNA obtained from the cDNA library screening reported by He et al. [2011].
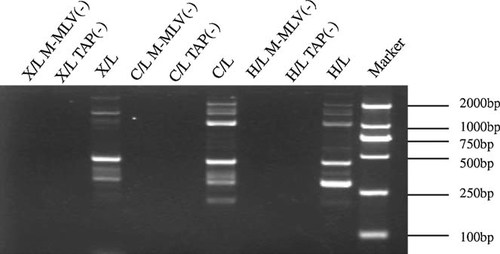
5′ RACE of UL145 of strains H, C, and X in L expression kinetics. H, C, and X strains had the same bands between 250–500 bp and 1,000–2,000 bp. The band at approximately 200 bp in the L RNA of C strain was an unspecific product. All the controls of TAP(−) and M-MLV(−) in three strains were negative.
The above results indicated that in addition to the polycistron identified by He et al. [2011], one monocistron containing only the sequence of UL145 ORF also existed, which could originate from multiple 5′ends. To obtain the full-length transcript, the sequences obtained from 5′ and 3′ RACE were linked together based on their overlapping sequences. The length of this monocistron was 500–700 bp, consistent with the results of the Northern blot.
All linked sequences have been submitted to Genbank: HQ537459, HQ537461-HQ537467, and HQ537469-HQ537474. The map of the ORFs and the transcripts are shown in Figure 4.
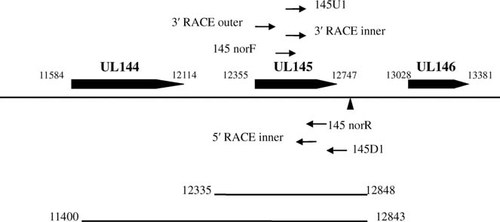
Map of the ORFs in the region under study. The positions are marked based on the DNA sequence of H strain (GenBank accession No. GQ981646). Two types of transcripts were obtained in this study. One contains UL145-specific sequence and the other contains both UL144- and UL145-specific sequences. The positions of 5′ termini of each primers corresponding to the H strain are listed in the table. Solid triangle denotes the position of poly (A) signals (AATAAA sequence) of transcripts obtained in the study, which locates at 12,821–12,826.
DISCUSSION
The HCMV UL145 gene is highly conserved in all strains isolated from patients, irrespective of whether the strains come from patients with different clinical features, or from different regions of the world. Phosphorylation motif sites for PKC and CK II in the putative UL145 protein of all strains are highly conserved. The relative conservation of UL145 sequences being retained under selection pressure suggests that it might be essential for virus survival and replication in vivo.
Qi et al. [2011] have identified the transcripts of the UL138-UL145 region as two large families of polycistronic transcripts, with the UL145 gene representing one component of the second family. He et al. [2011] have confirmed that UL144 and UL145 are expressed as a polycistron. In the current study, two abundant clusters of UL145 transcripts in H, C, and X clinical isolates were detected by Northern blot in late kinetics. Although the bands were smeared in a narrow area, the size of the transcripts was similar in the three strains isolated from patients. The cluster of transcripts between 1,400 and 1,600 nt in the Northern blot should represent transcripts from the polycistron of UL144 and UL145 identified by He et al. [2011]. However, one monocistron only containing the sequence of the UL145 ORF was confirmed in the present study by several evidences, including the results of cDNA library screening, Northern blot and RACE. Further, the 5′ RACE results of the three strains demonstrated that this monocistron could originate from multiple 5′ ends. In addition to transcribing with other genes, the UL145 gene had the ability of independent transcription. These results revealed the complex nature of the transcription of UL145 in HCMV strains. The transcriptional regulation under different originations is under current investigation.
The Northern blot results demonstrated that the transcript kinetics of the UL145 gene in the three strains were not in complete accord. To confirm the transcriptional kinetics of the UL145 gene in the three strains, Northern blots were repeated several times. Although the RNA extractions used in these experiments were not complete the same batch of cells, similar results were obtained for each strain in the repeated experiments. The possible upstream regulation sequence of the monocistron found in the present study should be located in the UL144 ORF, which has been demonstrated to be highly variable and divided into three groups [Lurain et al., 1999; Mao et al., 2007]. Sequence alignment was performed using the UL144 sequences of the three strains we have previously reported [He et al., 2011]. The identity scores among the three strains were only 78.8–85.3%. With previous analyses of the UL144 polymorphism [Lurain et al., 1999; Mao et al., 2007] as a reference, the UL144 sequences of H, C, and X strains should be assigned into different groups, respectively. Therefore, the varied regulation sequences upstream UL145 (in UL144) could lead to the discrepancy of the transcription kinetics of the UL145 gene among different strains.
Previous studies showed that the relative conservation of UL145 sequences was retained under selection pressure [Sun et al., 2007]. This suggests that it might be essential for virus survival and replication in vivo. Further investigation is warranted to determine if any significant differences exist among the transcription kinetics between different strains.




