Molecular epidemiology of measles virus in Iran 2009–2010: First detection of measles genotype H1
Abstract
Measles virus (MV) genotyping is an important component of measles surveillance in the context of monitoring immunization program effectiveness and documenting MV elimination. The molecular epidemiology and genetic variability of circulating MV strains in Iran during the 2009–2010 were studied in consecutive MV isolates from throat swab and urine. Sequence information obtained from 41 cases based on the 456 nucleotides of the most variable region of the C-terminal part of the N-protein revealed that these sequences belonged to two different genotypes. This is the first description of the genetic characterization of sporadic MV genotype H1 cases in northern Iran. Cases were probably linked to MV importation from distant parts of Asia. The genotype H1 has not been detected in the Eastern Mediterranean Region. In addition, both sequence analysis and epidemiologic data indicated that the more recently detected genotype D4 viruses in Iran were related very closely to viruses that were detected in Pakistan, suggesting that these viruses may have been imported from Pakistan. J. Med. Virol. 83:2200–2207, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Measles is a highly infectious respiratory infection caused by measles virus (MV). Despite widespread use of a safe and effective vaccine for over 40 years, measles remains endemic in many developing countries [Griffin, 2001]. Global mortality attributed to measles has been estimated to be 164,000 deaths in 2008 [CDC, 2009; WHO, 2009b]. Human MV is a member of the genus Morbillivirus of the Paramyxoviridae family [Griffin, 2001]. The N protein of wild-types viruses exhibits antigenic heterogeneity in the C-terminal region and differs up to 7–12% within the C-terminal 456 nucleotides [Rota et al., 1992; Xu et al., 1998]. Strains examined to date can be separated into 8 different clades and at least 23 different genotypes [CDC, 2005; WHO, 2005]. Using genotype information from 31 countries submitted to WHO genotype database seven separate measles genotypes were distinguished in 2007 (B3, D4, D5, D6, D8, D9, and H1). For example: D5 in Japan [Taira et al., 2008]; D9 in the Lao People's Democratic Republic, Malaysia and New Zealand; and H1 in Hong Kong and Vietnam [WHO, 2008, 2009a]. The WHO Regional Office for the Eastern Mediterranean (WHO/EMRO) aimed at elimination of measles in the region in 2010 [WHO, 2003; Anonymous, 2005]. Measles was an endemic disease in Iran and caused significant morbidity and regular outbreaks before initiation of the measles surveillance program in 1980 [Esteghamati et al., 2007]. Vaccination coverage in Iran has ranged from 38% to 99% since 1980 until 2005 with sustained high coverage (≥94–99%) during the past decade [Esteghamati et al., 2007].
The molecular epidemiology of circulating MV can be used to recognize transmission pathways of the virus. Molecular epidemiology enables enhanced understanding of epidemiological links during outbreaks and provides a means to measure the status of local measles surveillance programs [Bellini and Rota, 1998]. From all genotypes, genotype D4 has been detected continuously in Iran since 1998 and is therefore considered the indigenous genotype. D4 viruses have been reported regularly from different regions of Iran since mass vaccination in 2003 [Naseri et al., 2011]. After the measles/rubella mass vaccination campaign in 2003, the National Measles Laboratory of Iran has intensified molecular measles surveillance in remote geographic areas of the country. The National Measles Laboratory of Iran has investigated measles outbreaks in collaboration with Center for Disease Control [CDC, 2005] and Ministry of Health and Medical Education (MOHME) in Iran.
In the present study a genotype H1 outbreak is described by phylogenetic analysis of the N gene of MV. Sequence analysis confirmed that the D4 genotype remains the predominant circulating genotype since the introduction of MV mass vaccination in 2009–2010. Phylogenetic relationships were examined in order to explain the epidemiological links between different outbreaks.
MATERIALS AND METHODS
Specimens of blood, urine and throat swab from suspected cases of measles in 2009–2010 were collected from sentinel sites, hospitals, and outbreaks in different regions of Iran by CDC and MOHME in Iran. All the specimens were obtained according to WHO procedures [WHO, 1999, 2007] and transported to the National Measles Laboratory of Iran in the School of Public Health, Tehran University of Medical Science for processing. Detection of IgM specific antibodies to measles in sera was performed using a commercial ELISA kit (Dade Behring, Marburg, Germany).
Virus isolation was performed on urine and throat swabs of confirmed serologically measles cases. For virus isolation, the specimens were inoculated onto Vero/hSLAM cell line [Ono et al., 2001]. If virus culture was not successful for direct detection of MV, total nucleic acids were extracted from throat swabs and urine. MV RNA was extracted by the Nucleospin RNA Virus kits according to manufacturer's instructions (M&N Machery-Nagel, Dueren, Germany). Partial nucleoprotein gene segments of MV including the 456 terminal nucleotides were amplified by RT-PCR as described previously [CDC, 2005]. First round PCR products were sequenced in both directions using the ABI prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystems, Langen, Germany) by Genefanavaran Co. Wild-type measles isolates and genotype sequences from Iran were named as recommended by WHO [1998]. Genotype identification of the sequences was performed according to protocols by comparison to the WHO reference sequences, representative of the 23 established measles genotypes [WHO, 2001, 2007]. Sequenced data were analyzed by Clustal X [Thompson et al., 1997] and BioEdit version 7.0.0 DNA analysis software [Hall, 1999]. Phylogenetic trees were constructed using treecon package version 1.3b [Van de Peer and De Wachter, 1997] using the nucleotide Kimura-2 parameter and the neighbor-joining method. Bootstrap analyses were performed by 1,000 resampling of the data sets.
RESULTS
A total of 130 cases of measles in 2009 and 86 until May 2010 were confirmed by ELISA. Genetic analysis was undertaken on 41 confirmed measles cases (Table I). Thirteen sequences (31.7%) were obtained from viral isolates using Vero/hSLAM cell culture and the 28 (68.3%) were obtained directly from clinical samples. Clinical and demographic information for the 41 sequences are shown in Table I. The age of patients ranged from 4 months to 27 years and 83% were younger than 20 years. It was shown that 22% were vaccinated against MV (routine or campaign vaccination), 54% unvaccinated (12% of whom were not eligible for vaccination), and 24% had no vaccination history available (Table I).
| Strain designation | Age (year) | Sex | Days after rash onset | Source of specimen | Genotype | Province of origin | Comment | Vaccination history | Accession number |
|---|---|---|---|---|---|---|---|---|---|
| MVs/Zabol.IRA/17.09/1 | 3 | M | 1 | TSa | D4 | East | Small East outbreak | Yes | GQ504201 |
| MVs/Zabol.IRA/17.09/2 | 1 | M | 1 | TS | D4 | East | Small East outbreak | No | GQ504205 |
| MVs/Zabol.IRA/19.09 | 9.3 | F | 4 | TS | D4 | East | Small East outbreak | Yes | GQ504202 |
| MVs/Tehran.IRA/21.09/1 | 6.6 | M | 7 | TS | D4 | Center | Yes | GQ504199 | |
| MVs/Tehran.IRA/21.09/2 | 8 | F | 8 | TS | D4 | Center | Yes | GQ504204 | |
| MVs/Tehran.IRA/21.09/3 | 6.2 | M | 5 | TS | D4 | Center | No | GQ504203 | |
| MVs/Tehran.IRA/21.09/4 | 4.4 | M | 5 | TS | D4 | Center | No | GQ504200 | |
| MVs/Zahedan.IRA/22.09 | 7 m | M | 3 | TS | D4 | South-East | South-East outbreak | Unknown | GQ504198 |
| MVs/Zahedan.IRA/23.09 | 10 m | F | 6 | Urine | D4 | South-East | South-East outbreak | No | GQ504197 |
| MVs.Sistan.IRA/23.09/1 | 1.3 | M | 7 | TS | D4 | South-East | South-East outbreak | No | HM236421 |
| MVs.Tehran.IRA/25.09 | 4.5 | F | 4 | TS | D4 | Center | No | GQ504208 | |
| MVi/Savadkooh.IRA/25.09/1 | 5.5 | F | 1 | TS | D4 | North | Small north outbreak | Yes | GQ504210 |
| MVs/Babol.IRA/25.2009/2 | 19.8 | M | 3 | TS | D4 | North | Small north outbreak | No | GQ504213 |
| MVs.Savadkooh.IRA/26.09 | 1.4 | F | 3 | TS | D4 | North | Small north outbreak | No | HM236422 |
| MVs/Tehran.IRA/29.2009/1 | 5 | F | 4 | TS | D4 | Center | No | GQ504211 | |
| MVs/Tehran.IRA/29.2009/2 | 6 | F | 9 | TS | D4 | Center | Imported casesb | No | GQ504212 |
| MVs/Tehran.IRA/29.2009/3 | 3 | F | 5 | TS | D4 | Center | No | GQ504214 | |
| MVs.Tehran.IRA/29.09/4 | 4.4 | F | 9 | TS | D4 | Center | Imported casesb | No | HM236423 |
| MVs/Sari.IRA/30.2009/1 | 11m | M | 2 | TS | H1 | North | First reportc; small north outbreak | No | GQ504207 |
| MVs.Sari.IRA/30.2009/2 | 10 | F | 6 | TS | D4 | North | Small north outbreak | Unknown | GQ504209 |
| MVi.Sari.IRA/30.2009/3 | 5 | F | 5 | TS | H1 | North | First reportc; small north outbreak | No | GQ504206 |
| MVi.Sari.IRA/33.09 | 1.3 | M | 4 | TS | H1 | North | First reportc; small north outbreak | No | HM236424 |
| MVi.Sari.IRA/34.09 | 5.6 | F | 5 | TS | H1 | North | First reportc; small north outbreak | No | HM236425 |
| MVs.Karaj.IRA/42.09 | 5 | F | 4 | TS | D4 | Center | Unknown | HM236426 | |
| MVs.Tehran.IRA/47.09 | 20 | M | 4 | TS | D4 | Center | No | HM236427 | |
| MVs.Tehran.IRA/48.09/1 | 20 | M | 4 | TS | D4 | Center | No | HM236428 | |
| MVS/Varamin.IRA/48.09/2 | 23 | M | 5 | TS | D4 | Center | Unknown | HM236429 | |
| MVS/Isfahan.IRA/50.09 | 27 | M | 4 | TS | D4 | Center | Unknown | HM440230 | |
| MVS/Tehran.IRA/53.09/1 | 20 | M | 3 | TS | D4 | Center | Imported casesb | Unknown | HM440231 |
| MVS/Varamin.IRA/53.09/2 | 27 | M | 3 | TS | D4 | Center | Unknown | HM440232 | |
| MVi/Larestan.IRA/53.09/3 | 18 | F | 3 | TS | D4 | South | Unknown | HM440233 | |
| MVi/Sistan.IRA/03.10 | 1 | F | 5 | Urine | D4 | South-East | Yes | HM440225 | |
| MVs/Tehran.IRA/4.10/1 | 10m | F | 3 | TS | D4 | Center | Link to the Sought-East outbreak | No | HM002637 |
| MVi/Sistan.IRA/4.10/2 | 10 | M | 4 | Urine | D4 | South | Sought-East outbreak | Yes | HM002638 |
| MVi/Sistan.IRA/4.10/3 | 9 | M | 3 | Urine | D4 | South | Sought-East outbreak | Yes | HM002639 |
| MVi/Sistan.IRA/4.10/4 | 9 | M | 3 | Urine | D4 | South | Sought-East outbreak | Yes | HM002640 |
| MVs/Sistan.IRA/4.10/5 | 10 | F | 4 | Urine | D4 | South | Sought-East outbreak | No | HM002641 |
| MVi/Bandarabas.IRA/05.10/1 | 4 m | M | 4 | Urine | D4 | South | Sought outbreak | No | HM440226 |
| MVi/Bandarabas.IRA/05.10/2 | 9 m | F | 6 | Urine | D4 | South | Sought outbreak | No | HM440227 |
| MVi/Bandarlengeh.IRA/08.10/1 | 15 | F | 1 | Urine | D4 | South | Sought outbreak | Unknown | HM440228 |
| MVi/Bandarlengeh.IRA/08.10/2 | 27 | M | 3 | Urine | D4 | South | Sought outbreak | Unknown | HM440229 |
- a Throat swab.
- b Afghanis who are imported cases.
- c Small north outbreak presumably due to MV importations from Southeast Asian regions.
The derived sequences are recorded in GenBank (accession numbers: GQ504197–GQ504214, HM236421–HM236429, HM440226–HM440233, and HM002637–HM002641). Segregation of the Iranian strains into two distinct genotypes, D4 and H1, was revealed by BLAST software Clustal X after sequence analysis of C-terminal 456 nucleotides of the nucleoprotein (N) gene as recommended by WHO [2003]. Sequence analysis showed that, 37 of the 41 samples belonged to genotype D4 and 4 samples to genotype H1 (Table I, Fig. 1). This was the first time that an H1 MV was detected in Iran and the Eastern Mediterranean Region. None of the cases had any history of traveling to outside the country. BLAST analysis of partial sequences of the N gene of the Iranian H1 measles isolates confirmed they 100% were identical to the Taiwan and Hong Kong isolates, circulating during 2008 (MVs/Taoyuan.TWN/29.08 and MVs/Hong Kong.CHN/17.08) (Fig. 3). All four H1 isolates had sequence homology of 97.8% with the WHO reference sequence (Hunan.CHN/93/7[AF045212]) (Table II). A significant bootstrap value (100% for 1,000 replicates) confirmed clustering of the four Iranian MV strains within genotype H1 (Figs. 2 and 3). The Iranian H1 genotype was compared with different H1 sequences reported previously [Liffick et al., 2001; Yu et al., 2007]. The nucleotide and amino acid sequence homology with the H1a reference strain (China 93/2) was 98.9% and 97.3%; with the H1b reference strain (China 93/7) was 97.8% and 96%; with H1c reference strain (China 94/7) was 96.9% and 96% and with H2 reference strain (China 94/1) was 92.9% and 91.3% (Table II). Phylogenetic analysis (Fig. 3) indicated that all four study isolates were closest related to the H1a reference strain (China 93/2). The nucleoprotein C terminus of the H1 Iranian strains showed four amino acid differences with the H1a reference strain (G422S, S457G, L475I, and R497K). In addition, the H1a reference strain (China 93/2) and all four Iranian strains had E at amino acid 484 [Yu et al., 2007]. The average nucleotide difference between the Iranian H1 and D4 strains and the MV vaccine strain Edmonston B nucleoprotein gene (U03656) belonging to genotype A exhibited a 7.1% and a 6.7% divergence, respectively.
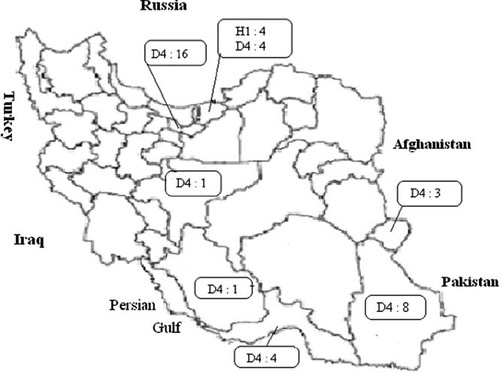
Geographical distribution of measles virus genotypes in Iran (2009–2010). Numbers and type of genotype are shown in boxes.
| Seq→ | IRA/30.09/1 | IRA/30.09/3 | IRA/33.09 | IRA/34.09 | CHN/93/2(H1a) | CHN/94/7(H1c) | CHN/93/7(H1b) | CAN/8.01/1(H1) | MOG/50.02(H1) | CHN/94/1 (H2) |
|---|---|---|---|---|---|---|---|---|---|---|
| IRA/30.09/1 | ID | 100 | 100 | 100 | 98.9 | 96.9 | 97.8 | 98.6 | 99.5 | 92.9 |
| IRA/30.09/3 | 100 | ID | 100 | 100 | 98.9 | 96.9 | 97.8 | 98.6 | 99.5 | 92.9 |
| IRA/33.09 | 100 | 100 | ID | 100 | 98.9 | 96.9 | 97.8 | 98.6 | 99.5 | 92.9 |
| IRA/34.09 | 100 | 100 | 100 | ID | 98.9 | 96.9 | 97.8 | 98.6 | 99.5 | 92.9 |
| CHN/93/2(H1a) | 97.3 | 97.3 | 97.3 | 97.3 | ID | 97.5 | 98.4 | 99.3 | 99.3 | 93.6 |
| CHN/94/7(H1c) | 96 | 96 | 96 | 96 | 97.3 | ID | 98.2 | 97.3 | 97.3 | 94.7 |
| CHN/93/7(H1b) | 96 | 96 | 96 | 96 | 97.3 | 97.3 | ID | 98.2 | 98.2 | 94.5 |
| CAN/8.01/1(H1) | 98 | 98 | 98 | 98 | 99.3 | 98 | 98 | ID | 99.1 | 93.4 |
| MOG/50.02(H1) | 99.3 | 99.3 | 99.3 | 99.3 | 98 | 96.6 | 96.6 | 98.6 | ID | 93.4 |
| CHN/94/1(H2) | 91.3 | 91.3 | 91.3 | 91.3 | 92 | 92.7 | 93.3 | 92.7 | 91.3 | ID |
- Nucleotide sequence identity is in the right upper corner; amino acid sequence identity is in the left lower corner. IRA/30.09/1, IRA/30.09/3, IRA/33.09, IRA/34.09 measles isolated from this study, see Table I.
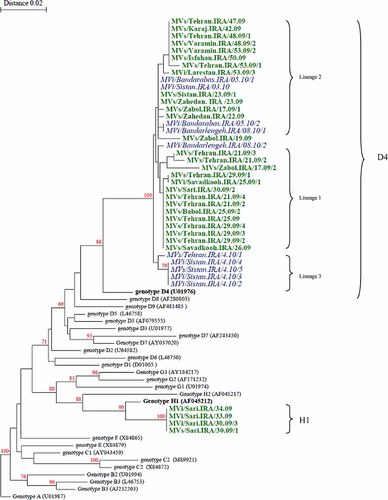
Phylogenetic tree of the C-terminus of N gene sequences of 41 wild-type measles isolates from Iran compared to the WHO reference sequences for each genotype. The Iranian sequences were isolated in 2009 are shown in bold (D4 and H1 genotypes). The Iranian sequences were isolated in 2010 are shown in italics (D4 genotype). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
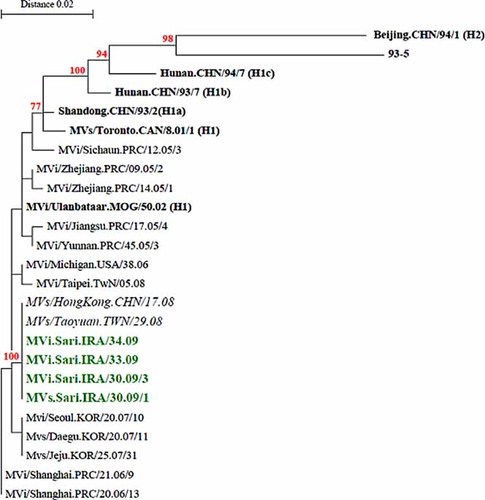
Phylogenetic comparison of four Iranian measles virus strains (C-terminus of N gene) isolated in 2009 with the H1 reference sequences and other wild-type measles genotype H1 detected in other countries. Bootstrap values (1,000 replicates) >70% are indicated. The Iranian sequences are shown in bold. The H1 reference sequences are shown in black and bold. Note: The closely related sequences (MVs/Taoyuan.TWN/29.08 and MVs/HongKong.CHN/17.08) with the highest nucleotide BLAST scores from GenBank (100% identity) are shown in italics. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Nine distinct lineages of MV genotype D4 were reported in Iran [Naseri et al., 2011]. The average genetic diversity of genotype D4 before measles mass vaccination (2003) was significantly higher than recent years (2.2% vs. 0.9%) [Naseri et al., 2011]. In the present study there were at least three separate lineages of genotype D4. Sequences which were identical or very similar are referred to as lineages 1–3 (Fig. 2). Some of the Iranian strains in 2010 had similar sequences (99.5–100% identity) to genotype D4 measles obtained in 2009 (Fig. 2). NCBI blast analysis illustrated that the nucleotide sequences of Iranian D4 strains MVs/Zahedan.IRA/23.09, MVs.Sistan.IRA/23.09/1, MVi/Sistan.IRA/03.10, and MVi/Bandarabas.IRA/05.10/1 were identical to Iranian strains described in 2008 (MVs/Khorasan.IRA/20.08 and MVi/Mazandaran.Iran/10.08) [Naseri et al., 2011] as well as the MVs/Mandi Bahuddin.PAK/426.07 (FJ790762) Pakistanian strain which was reported in 2007 (Fig. 4).

The Iranian sequences were isolated in 2009 are shown in bold. The Iranian sequences were isolated in 2010 are shown in italics. The D4 reference sequence is shown as a root. Note: The closely related sequences from Pakistan with the highest nucleotide BLAST scores from GenBank (99–100% identity) are shown in bold. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The Iranian D4 strains belonged to lineage 1 (HM236427–HM236429, HM440225, HM440230–HM440233) and lineage 2 (GQ504208–GQ504214, HM236422, HM236423, GQ504199, GQ504200) are more closely related (99% identity) to wild-type MVes from Pakistan which were circulating in 2007 and probably continued to circulate during 2009 (MVs/Mandi Bahuddin.PAK/426.07). In addition, the Iranian D4 strains which belonged to lineage 3, MVs/Tehran.IRA/4.10/1, MVi/Sistan.IRA/4.10/2, MVi/Sistan.IRA/4.10/3, MVi/Sistan.IRA/4.10/4, MVs/Sistan.IRA/4.10/5 (HM002637–HM002641) had 99% identity to sporadic case (MVs/Ontario.CAN/15.08) reported by Tipples et al. in 2008 in GenBank which are both closely related to Pakistanian strain (FJ790762) (Fig. 4).
The majority of the D4 isolates in 2009–2010 were associated with small outbreaks in different parts of Iran, whereas the H1 genotypes were associated with a small outbreak in northern Iran (Figs. 1 and 2). Except African and Eastern Mediterranean Regions, all WHO regions have now reported the circulation of MV genotype H1.
DISCUSSION
Historically, measles and its complications have been an important public health challenge for Iran. Despite the fact that measles vaccination was initiated in 1967, regular outbreaks of measles have been reported since the early 1980s, which continued until 2002 [Esteghamati et al., 2007]. Iran has made progress in measles control and elimination by the establishment of keep-up, catch-up, follow-up, and mop-up programs [Esteghamati et al., 2007]. After the measles/rubella mass vaccination campaign in 2003, the measles vaccination coverage increased to 98% but small outbreaks or sporadic cases still occurred [Esteghamati et al., 2007; Naseri et al., 2011]. In this study, phylogenetic analysis of partial N gene nucleotide sequences of Iranian measles isolates were compared with published N gene sequences and WHO reference strains. Four of the Iranian strains clustered with genotype H1 which were all related to a small outbreak in northern Iran in 2009. In addition, the nucleotide sequence and amino acid homology between Iranian H1 strains was 100% (Table II). Although the source of Iranian H1 strains were not identified in this study, the introduction of H1 was probably due to importations from Southeast Asian regions, as they are identical to genotype H1 from Taiwan (MVs/Taoyuan.TWN/29.08 [GQ338674]). Sequence homology was 100% at the nucleotide level of hypervariable region of the N gene sequences [Cheng et al., 2009].
The patients infected with MV H1 had no history of travel abroad and their transmission pathway could not be traced. All cases were difficult-to-reach populations such as nomadic communities with low coverage of MV vaccination. All patients may have been infected by road workers from Southeast Asian regions. Sequence analysis of wild-type H1 MV isolated in countries in Asia has confirmed that endemic circulation of genotype H1 is limited mainly to China [Rota et al., 2009]. Phylogenetic analyses conducted during the early part of the last decade identified two clusters (cluster 1 and 2) within the Chinese genotype H1 [Zhang et al., 2007, 2009]. Previously, importation of genotype H1 viruses into Taiwan, Hong Kong, and Europe, for which 100% sequence homology was shown with genotype H1 viruses which were circulating in mainland China [Ji et al., 2010]. This is in line with the present study in which phylogenetic analysis showed that the Iranian H1 strains were members of Cluster 1. These four isolates were closely related to the H1a reference strain (China 93/2).
The results of this study indicate that the majority of MVs collected between 2009 and 2010 belonged to genotype D4 (37 isolates), which is the dominant strain in Iran. The phylogenetic and sequence analysis of D4 measles strains before mass vaccination in Iran exhibited much more viral complexity than the current D4 strains. Identical sequences were detected in multiple distant provinces at the same time. After mass vaccination the number of co-circulating chains of transmission has clearly been reduced. In the current study, the average genetic diversity of the Iranian D4 genotype was limited to 0.9% suggesting that a single introduction of virus was responsible for the outbreaks. This is similar to a report from Burkina Faso in 2003 [Mulders et al., 2003]. During the same period, genotype information of the neighboring countries of Iran indicated that there were different measles genotypes, such as D4, D5, D6, D8, H1 in Russia [Astrasheuskaya et al., 2008; Shulga et al., 2009], D4 in Iraq, Syria, Pakistan, and Afghanistan [Riddell et al., 2005], D6, D4 in Turkey [Korukluoglu et al., 2005]. NCBI blast analysis showed that the D4 Iranian strains belong to the three lineages so that most outbreaks and sporadic cases in 2009 were due to lineages 1 and 2 whereas linage 3 was found predominantly in 2010 (Fig. 2). The Iranian viruses were related very closely to viruses which were detected in Pakistan, suggesting that these viruses could have been imported from Pakistan in 2008 and continued to circulate in southern Iran since it was detected in 2009 (Fig. 4).
Because of high vaccination coverage in Iran, the transmission chain of several indigenous MV strains has been interrupted. However, it is difficult to document interruption of transmission of the suspected indigenous genotype because genotype D4 is being reintroduced by continuously importation from neighboring countries and may cause eventually a massive spread of measles throughout much of the susceptible region [Edmonson et al., 1990]. In regions which have eliminated wild-type indigenous measles, importation of measles is now the main cause of outbreaks. In this study patients with measles resided close to neighboring countries where measles outbreaks are common.
Iran is one of the countries in the Eastern Mediterranean Region which conducts routine molecular surveillance of MV associated with sporadic cases and outbreaks. Although mass vaccination has decreased successfully the incidence of the disease, outbreaks continue to occur. Therefore, in order to reach the goal set by WHO to eliminate measles from the Eastern Mediterranean Region by 2010 [WHO, 2003; Anonymous, 2005], increased active surveillance and molecular characterization of the strains is crucial for monitoring the progress in measles elimination.
In conclusion, the present study is the first description of an H1 MV outbreak outside Taiwan in the Eastern Mediterranean Region. It highlights the necessity of molecular MV surveillance for rapid detection of new MV strains. Iran had attained to the main criteria (achieving and maintaining at least 95% vaccination coverage annually and 90% reduction in mortality and morbidity) for interruption of indigenous measles transmission but could not interrupt completely the transmission of measles by 2010. Iran is heading towards elimination of MV by enhanced molecular MV surveillance and according to the new measles elimination strategic plan in the Eastern Mediterranean Region, Iran like other countries in this region will hopefully eliminate the indigenous virus by 2015.
Acknowledgements
We thank the staff of the Virology Division, School of Public Health, Tehran University of Medical Sciences, and Center for Diseases Management in the Ministry of Health, Iran for assistance in collection of data and samples.




