Characterization of norovirus strains in Australian children from 2006 to 2008: Prevalence of recombinant strains†
None of the authors have conflicts of interest.
Abstract
Noroviruses are highly infectious and are the most common cause of gastroenteritis outbreaks. Genotype II.4 strains have been the dominant type identified in adults, however the genotype distribution in children is less clearly defined. This study aimed to detect and genotype norovirus strains infecting children hospitalized with acute gastroenteritis in Melbourne, Australia from 2006 to 2008. Stool samples were collected from 272 children admitted to the Royal Children's Hospital, Melbourne, Australia, with non-rotavirus acute gastroenteritis between April 2006 and December 2008. Using RT-PCR, norovirus was detected in 36% of samples. Strains were genetically characterized via analysis of regions from both the capsid gene and the RNA dependent RNA polymerase (RdRp) gene, to investigate genotype distribution and incidence of recombination. Typing based on the capsid gene (n = 70) detected GII.4 (49%) and GII.3 (46%) as the most predominant genotypes. Strains with a GII.4 capsid were usually assigned a GII.4 RdRp, whereas most strains identified as GII.3 based on capsid typing were assigned a GIIb RdRp (71%). The GII.3/GIIb represent recombinant strains. Sequence analysis of the putative recombination breakpoint was performed for three representative suspected recombinants: GII.3/GIIb (n = 2) and GII.3/GII.12 (n = 1). Recombination analysis confirmed these strains as recombinants and identified putative breakpoints adjacent to the ORF1/ORF2 junction. This study highlights the importance of norovirus infection as a cause of pediatric gastroenteritis. It also reinforces the high circulation of recombinant strains causing disease in children, particularly the GII.3/GIIb strain. J. Med. Virol. 83:2213–2219, 2011. © 2011 Wiley Periodicals, Inc.
INTRODUCTION
Norovirus is the most common cause of gastroenteritis in people of all ages [Mead et al., 1999] and is estimated to cause 50% of all gastroenteritis outbreaks worldwide [Patel et al., 2008]. It is highly contagious, with transmission occurring mostly through person to person contact or ingestion of contaminated food or water [Becker et al., 2000].
The genus Norovirus is a member of the family Caliciviridae, and contains viruses with a positive sense, single stranded RNA genome of approximately 7.5 kb [Jiang et al., 1990; Green et al., 2000]. The genome is organized into three open-reading frames (ORF). ORF1 encodes the non-structural proteins, including the RNA-dependent RNA polymerase (RdRp); ORF2 encodes the capsid protein (VP1) and ORF3 encodes a minor structural protein (VP2) [Jiang et al., 1993; Bertolotti-Ciarlet et al., 2003].
Noroviruses are classified into five genogroups based on sequence analysis of regions within the capsid and RdRp gene. Human noroviruses are classified into three of these genogroups: GI, GII, and GIV, with the majority of human strains belonging to the GII group. Human noroviruses can be further divided into at least 25 different genotypes [Zheng et al., 2006]. Genotype II.4 (GII.4) is the most common worldwide and has caused at least four global epidemics since 1995, each caused by a distinct GII.4 antigenic variant [Noel et al., 1999; Siebenga et al., 2009]. A new “2008 variant” has been reported in many countries since 2008, and may represent the latest epidemic variant [Belliot et al., 2010; Kremer et al., 2011; Mans et al., 2010; Siebenga et al., 2010].
Sequence analysis of both the capsid and RdRp genes, has detected the circulation of recombinant norovirus strains. For most norovirus recombinants, the breakpoint occurs adjacent to the 20 bp ORF1/ORF2 overlap region. Recombinant strains have been widely reported in recent years, and a number of novel RdRp clusters which have no corresponding capsid type have been identified [Bull et al., 2007].
Following rotavirus, norovirus is the second most important cause of gastroenteritis in children, leading to more than 200,000 deaths annually [Patel et al., 2008]. A previous Australian study characterized norovirus strains collected from 1998 to 2002 from pediatric patients hospitalized with gastroenteritis. A prevalence of 9% was determined and genetic analysis of the RdRp region found GII.4 to be predominant [Kirkwood et al., 2005]. Recent studies have reported an increased diversity of genotypes infecting children compared to those isolated from adults, as well as a frequent detection of recombinant strains [Oh et al., 2003; Lindell et al., 2005; Phan et al., 2007; Beersma et al., 2009; Chhabra et al., 2009; Jin et al., 2009].
The aim of the current study was to determine the prevalence and genotype of norovirus strains in children hospitalized with acute gastroenteritis in Melbourne, Victoria, Australia, from 2006 to 2008.
MATERIALS AND METHODS
Specimen Collection
A total of 272 stool specimens were collected from children admitted to the Royal Children's Hospital, Melbourne, Australia, with acute gastroenteritis, between April 2006 and December 2008, for norovirus detection. All specimens were determined as rotavirus-negative by commercial ELISA. An additional ten strains collected from children in Melbourne from 2002 to 2005 were also analyzed for a wider phylogenetic comparison.
RNA Extraction
Viral RNA was extracted from 20% fecal suspensions using the QIAamp viral RNA Minikit (Qiagen, Hilden, Germany).
RT-PCR Amplification of Regions of the Capsid and RdRp Genes
Norovirus was detected using a nested RT-PCR targeted to a region of the capsid gene as described previously [Bull et al., 2006]. First and second round primers were NV2oF2/NV2oR [Bull et al., 2006] and G2F3/G2SKR [Kojima et al., 2002; Hansman et al., 2004] which amplified a 266 bp region at the 5′ end of the capsid gene.
Two RT-PCR protocols were used to amplify distinct regions of the RdRp gene. Region A (274 bp) was amplified using primers P289/P290 [Jiang et al., 1999] and region B (172 bp) was amplified using primers Mon 431, Mon 432, Mon 433, and Mon 434 [Fankhauser et al., 2002]. Each method was performed as described previously [Jiang et al., 1999; Fankhauser et al., 2002; Vinje et al., 2004].
Amplification Across the Putative Recombination Breakpoint
To confirm recombination, a 1,049 bp region spanning the ORF1/ORF2 overlap was amplified using the RT-PCR superscript III kit (Invitrogen, Carlsbad, CA) and primers P290 and G2SKR. The amplified region corresponded to nucleotides 4,318–5,366 on the Lordsdale virus genome (accession no. X86557) and included 787 bp of ORF1 and 281 bp of ORF2.
All amplified cDNA products were electrophoresed on 1–1.5% agarose gels containing ethidium bromide and visualized under UV light.
Sequence Analysis of cDNA Products
Capsid and RdRp PCR amplicons were sequenced to perform genotype determination, while the 1,049 bp breakpoint amplicons were sequenced to determine breakpoint locations.
Amplified cDNA products were gel purified using the QIAquick Gel Extraction Kit (Qiagen) and sequenced in both directions using Big Dye Terminator v3.1 on an ABI 3130xl Genetic Analyser (Applied Genetic Diagnostics, Department of Pathology, University of Melbourne). Sequence data was analyzed using Sequencher version 4.1 (Gene Codes Corp., Ann Arbor, MI). JModelTest was used for model selection [Posada, 2008], and distances computed using the Kimura 2-parameter method [Kimura, 1980]. Blastn analyses were conducted [Altschul et al., 1997], together with alignments and phylogenetic analyses using MEGA version 4 [Tamura et al., 2007]. Evolutionary history was inferred using the neighbor-joining method [Saitou and Nei, 1987] with 1,000 bootstrap replicates [Felsenstein, 1985]. Genotypes were assigned as defined by Zheng et al. [2006].
Breakpoint Analysis
Recombination and putative breakpoint analysis was conducted using Simplot for Windows version 3.5.1 [Lole et al., 1999] and the maximum chi-squared test [Smith, 1992], calculated using START2 [Jolley et al., 2001]. Results were confirmed using phylogenetic analysis (MEGA 4) [Tamura et al., 2007]. Potential parent strains were Mexico virus (GII.3) (HCU22498), Saitama U1 (GII.12) (AB039775), and Hawaii virus (GII.1) (U07611). Picton virus (GII.1/GIIb) (AY919139) was used for phylogenetic analysis of GII.3/GIIb strains in the RdRp region and was omitted from Simplot and statistical tests as it is a recombinant strain.
RESULTS
Detection of Norovirus in Hospitalized Children With Acute Gastroenteritis
Between April 2006 and December 2008, 272 rotavirus-negative stool samples were collected from children admitted to the Royal Children's Hospital, Melbourne, Australia, with acute gastroenteritis. Norovirus was identified in 86 samples (36%). Its prevalence peaked in 2006, detected in 50% of samples; and norovirus was detected in 28% of 2007 and 2008 samples.
The majority of norovirus infections occurred in males (67%). Norovirus infections occurred in children aged 2 weeks to 16.7 years (average 3.8 years), with 49% from infants 12 months or younger (Table I).
| Age | ||||||
|---|---|---|---|---|---|---|
| ≤6 months | 7–12 months | 1–2 years | 2–5 years | 5–10 years | 10–17 years | |
| Proportion of positive isolates (%) | 26.5 | 22.9 | 14.45 | 10.84 | 8.43 | 15.66 |
Genotype Analysis of Norovirus Strains
Sequence data were obtained for 70/86 norovirus positive specimens. Each was genotyped based on genetic analysis of a 266 bp region of the capsid VP1 gene. All strains belonged to the GII genogroup, and the majority clustered with GII.4 (49%) and GII.3 (46%). As single strains, GII.1, GII.7, GII.13, and a novel type, which did not cluster with any standard strain, were identified (Fig. 1, Table II). The single novel strain shared high nucleotide identity (98–99%) with eight strains reported to GenBank, isolated from multiple countries from 2006 to 2009. This group formed a distinct cluster, termed GII.x (Fig. 1).
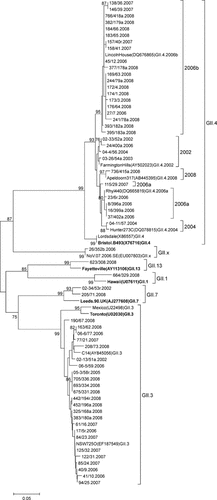
Phylogenetic analysis of a 266 bp region at the 5′ end of the capsid gene of norovirus strains. The year each strain was isolated is indicated in the strain name. Reference strains were selected from GenBank and the accession number and genotype is indicated. Genbank accession numbers JN602261 to JN602338 were assigned to capsid sequence determined in this study. The prototype strains defined by Zheng et al. [2006], used to classify genotype, are bolded. The tree is drawn to scale, with branch lengths in the units of the number of base substitutions per site. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches where the value is 70% or higher.
| Year | Samples tested | Positives | Strains sequenced | Genotype—cluster | No. of strains |
|---|---|---|---|---|---|
| 2006 | 38 | 19 | 17 | GII.4 | 10 |
| 2006a | 7 | ||||
| 2006b | 2 | ||||
| 2002 | 1 | ||||
| GII.3 | 6 | ||||
| GII.x | 1 | ||||
| 2007 | 63 | 18 | 16 | GII.3 | 9 |
| GII.4 | 7 | ||||
| 2006b | 6 | ||||
| 2006a | 1 | ||||
| 2008 | 171 | 49 | 37 | GII.3 | 17 |
| GII.4 | 17 | ||||
| 2006b | 16 | ||||
| 2008 | 1 | ||||
| GII.1 | 1 | ||||
| GII.7 | 1 | ||||
| GII.13 | 1 | ||||
| Total | 272 | 86 | 70 | GII.4 | 34 |
| 2006b | 24 | ||||
| 2006a | 8 | ||||
| 2002 | 1 | ||||
| 2008 | 1 | ||||
| GII.3 | 32 | ||||
| GII.1 | 1 | ||||
| GII.7 | 1 | ||||
| GII.13 | 1 | ||||
| GII.x | 1 |
Genotype predominance fluctuated between GII.3 and GII.4. GII.4 was dominant in 2006 (GII.4 59%, GII.3 35%), GII.3 was dominant in 2007 (GII.3 56%, GII.4 44%), and both genotypes were detected equally in 2008 (each 46%).
Five GII.4 variants: the 2002, 2004, 2006a, 2006b, and newly recognized 2008 variant were identified during this study, with the 2006b variant being predominant (71%). The 2004 variant cluster contained only a 2004 strain and the 2002 cluster contained strains from 2002 to 2004 and one 2006 isolate, indicating that GII.4 strains have continually evolved into new variants. Nucleotide similarity within the GII.4 cluster ranged from 94% to 100%. The GII.3 cluster exhibited a 96–100% nucleotide similarity. GII.3 strains from 2002 to 2005 clustered within the sequences from 2006 to 2008, indicating that similar GII.3 strains had been circulating at low levels in earlier years.
Capsid and RdRp Type Comparisons
Genotype assignment based on the RdRp gene was successful in 56% of samples analyzed (39/70). Phylogenetic trees for regions A and B of the RdRp gene are shown in Figure 2. The capsid and RdRp typing was concordant for 84% of capsid typed GII.4 strains, whereas only a single strain had both a GII.3 capsid and GII.3 RdRp. Strains with a GII.3 capsid were mostly detected in combination with the RdRp genotype IIb (71%) or GII.12 (12%), suggesting a high prevalence of recombinant strains. Singularly observed capsid/RdRp combinations included GII.4/GIIb, GII.4/GI.4, GII.3/sapovirus, GII.3/GII.4, GII.7/GII.7, GII.13/GIIb, and GII.x/GII.x (Table III). The intergenogroup and intergenus combinations are more likely to be mixed infections than recombinants.
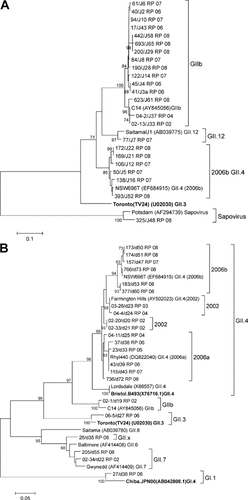
Phylogenetic analysis of (A) region A (274 bp) and (B) region B (172 bp) of the RdRp gene of norovirus strains. The year each strain was isolated is indicated in the strain name. Reference strains were selected from GenBank and the accession number and genotype is indicated. GenBank accession numbers assigned to Region A sequences determined in this study were JN602339 to JN602364. The prototype strains defined by Zheng et al. [2006], which were used to classify genotype where possible, are bolded. The tree is drawn to scale, with branch lengths in the units of the number of base substitutions per site. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches where the value is 70% or higher.
| Year | Genotypea capsid/RdRp | No. of strains |
|---|---|---|
| 2006 | GII.3/GIIb | 3 |
| GII.4/GII.4 | 3 | |
| GII.4/GIIb | 1 | |
| GII.3/GII.3 | 1 | |
| GII.4/GI.4 | 1 | |
| GII.x/GII.x | 1 | |
| 2007 | GII.3/GIIb | 4 |
| GII.4/GII.4 | 4 | |
| GII.3/GII.4 | 1 | |
| GII.3/GII.12 | 1 | |
| 2008 | GII.4/GII.4 | 10 |
| GII.3/GIIb | 5 | |
| GII.3/GII.12 | 1 | |
| GII.3/sapovirus | 1 | |
| GII.7/GII.7 | 1 | |
| GII.13/GIIb | 1 | |
| Total | GII.4/GII.4 | 17 |
| GII.3/GIIb | 12 | |
| GII.3/GII.12 | 2 | |
| GII.3/GII.4 | 1 | |
| GII.3/GII.3 | 1 | |
| GII.7/GII.7 | 1 | |
| GII.4/GIIb | 1 | |
| GII.13/GIIb | 1 | |
| GII.4/GI.4 | 1 | |
| GII.3/sapovirus | 1 | |
| GII.x/GII.x | 1 |
- a Capsid and RdRp genotype assignment was based on sequence data obtained from separate amplicons from the same stool specimen. Where the capsid and RdRp type is different, this may indicate a single recombinant strain or dual norovirus infection.
Overall the most common combinations observed were GII.4/GII.4 (44%), and the suspected recombinants GII.3/GIIb (31%) and GII.3/GII.12 (5%). GII.3/GIIb and GII.4/GII.4 were equally prevalent during 2006 (each 30%) and 2007 (each 40%). GII.4/GII.4 became more prevalent in 2008 (53%) compared to GII.3/GIIb (26%).
Recombination Analysis
Three potential recombinant strains (2 GII.3/GIIb, 1 GII.3/GII.12) were selected to investigate the presence of recombination and locate putative breakpoints.
Phylogenetic analysis was conducted for regions immediately upstream and downstream of the ORF1/ORF2 overlap (data not shown). The three samples clustered with Mexico virus in ORF2. Conversely, ORF1 clustered with either Picton virus (GIIb RdRp) for GII.3/GIIb strains or Saitama U1 (GII.12) for the GII.3/GII.12 strain. Simplot analysis supported this finding, with the capsid region of all simplots sharing >90% similarity to Mexico virus and the RdRp region showing high similarity with the alternate parent strain (Saitama U1 for GII.3/GII.12 and Hawaii for GII.3/GIIb). The GII.3/GIIb simplots were identical and all simplots had a visible cross-over event as shown in Figure 3.
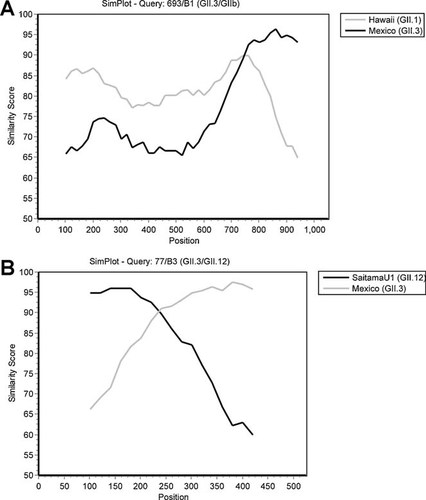
Similarity plots for strains (A) 693/B1 (GII.3/GIIb) and (B) 77/B3 (GII.3/GII.12). Plots were generated using the 2-parameter (Kimura) distance model [Kimura, 1980] in a sliding window of 200 nucleotides and a step size of 20. The nucleotide identity of suspected recombinants with each parental strain, (part A) Hawaii virus (GII.1) and Mexico virus (GII.3); or (part B) Saitama U1 (GII.12), and Mexico virus (GII.3), is plotted as a percentage for each nucleotide position. The 1,049 nucleotide positions plotted in part A correspond to nucleotides 4,318–5,366 on Lordsdale virus. The 525 nucleotide positions plotted in part B correspond to nucleotides 4,842–5,366 on Lordsdale virus. The predicted recombination breakpoint is indicated by the point where both parental strains share equal identity to the recombinant strain (where the two graphs cross). GenBank accession numbers JN602258, JN602259 and JN602260 were assigned to the three breakpoint sequences.
Based on the maximum chi-squared test, the three strains were verified as recombinants with breakpoints occurring adjacent to the ORF1/ORF2 overlap (P < 0.0001). The GII.3/GIIb strains had identical breakpoints, at nucleotide position 5,108 (Lorsdale virus numbering), located 25 bp from the GII.3/GII.12 breakpoint (5,133).
Discussion
Norovirus is an important agent causing severe acute gastroenteritis in children in Melbourne, with an average detection rate of 36% in rotavirus-negative stool specimens collected between April 2006 and December 2008. Incidence peaked in 2006 which was consistent with previous Australian [Tu et al., 2008] and international studies [Siebenga et al., 2009] where large outbreaks in adults and children were reported.
GII.4 was the most common genotype identified throughout the study period, similar to previous Australian reports of pediatric disease [Kirkwood et al., 2005]. The high detection rate of GII.3 strains identified in this study supports international findings that GII.3 is highly prevalent in pediatric settings [Nguyen et al., 2008; Chhabra et al., 2009; Jin et al., 2009].
The current study illustrated a yearly fluctuation in GII.3 and GII.4 strains. GII.4 was the most prevalent genotype in 2006, and subtype 2006a the most common variant, which is similar to that reported in outbreaks in adults in Australia and New Zealand [Tu et al., 2008]. Despite the overwhelming prevalence of GII.4 in adults in Victoria in 2006, GII.3 was responsible for a large portion of sporadic pediatric infections in this period. The relevance of GII.3 pediatric disease during this period was also evident in Chinese, Japanese, and Vietnamese pediatric studies, where GII.3 was dominant during the 2005–2006 norovirus season [Phan et al., 2007; Nguyen et al., 2008; Jin et al., 2009].
In this study GII.3 was dominant in 2007, as also observed in 2006–2007 in Indian children [Chhabra et al., 2009]. GII.3 and GII.4 were detected equally in 2008, together causing over 90% of norovirus disease. A dominance of these genotypes was also observed in sporadic pediatric cases in South Korea in 2008 [Chung et al., 2010]. Conversely, outbreak studies in adult populations in Australia, Europe and Canada reported the GII.4 2006b subtype as the dominant type in 2008, with little GII.3 activity [Siebenga et al., 2008; Eden et al., 2010; Pang et al., 2010]. Interestingly, 16/17 GII.4 strains detected in the current study in 2008, clustered with the 2006b variant. A single GII.4 2008 variant was identified; whether this variant will emerge globally remains to be seen.
A high proportion of recombinant strains, particularly GII.3/GIIb, were detected in this study. The reason for this is unknown, but perhaps a GIIb or GII.12 RdRp confers increased viral fitness compared to the GII.3 RdRp. GII.3/GIIb is emerging as an important pediatric genotype, having also been found in children in Japan in 2005–2006, India in 2006–2007, Egypt in 2006–2007, and Tunisia in 2003–2007 [Phan et al., 2007; Chhabra et al., 2009; Kamel et al., 2009; Sdiri-Loulizi et al., 2009]. Studies exploring RdRp type only, reported GIIb as the dominant pediatric genotype in the Netherlands in 2002–2007 [Beersma et al., 2009], Sweden in 2000–2003 [Lindell et al., 2005], and Germany in 2001–2002 [Oh et al., 2003].
A clear association of GIIb with pediatric disease was reported in a Swedish study, where simultaneous comparison of adult and pediatric disease cohorts, identified a much higher prevalence of GIIb in children [Lindell et al., 2005]. A French study observed a similar trend where 58% of GIIb strains were detected in children [Bon et al., 2005]. In Victoria, Australia, a quarter of GIIb outbreaks in 2002–2005 occurred in children. Furthermore, all pediatric GIIb outbreaks were caused by strains with a GII.3 capsid, whilst adult GIIb outbreaks were generally associated with a GII.1 capsid [Bruggink and Marshall, 2009].
This study confirms that norovirus is a common cause of acute gastroenteritis in Australian children. Whilst GII.4 is the major genotype causing outbreaks in the general population, other genotypes, especially the recombinant strain GII.3/GIIb, are also highly prevalent in pediatric disease. Pediatric norovirus strains vary considerably from those infecting adults, and a vaccine which intends to protect children from norovirus disease would need to incorporate a range of genotypes. Thus, continued surveillance is vital to appreciate the diversity of genotypes infecting children and adults.




