Mutations in the E2 and NS5A regions in patients infected with hepatitis C virus genotype 1a and their correlation with response to treatment†
Conflicts of Interest: None declared.
Abstract
Heterogeneity of subgenomic regions of hepatitis C virus (HCV) may be associated with response to interferon (IFN) therapy. The amino acid sequences of the PKR/eIF-2α phosphorylation homology domain (pePHD), IFN sensitivity determining region (ISDR), PKR binding domain (PKRBD), and variable region 3 (V3) were studied in 19 patients before and after 4 weeks of treatment. All patients were infected with HCV genotype 1a and were treated with pegylated-IFN and ribavirin. Thirteen patients achieved sustained viral response (responders) and six failed to clear viral RNA (nonresponders). The amino acid sequences in the pePHD and ISDR were identical in responders and nonresponders. However, amino acid substitution at position 2252 of PKRBD was significantly different between responders and nonresponders (P = 0.044). A larger number of mutations were observed in the V3 region of responders (P < 0.001). In this region, the amino acid in position 2364 differed between responders and nonresponders (responders: aspartic acid and serine, nonresponders: asparagine, P = 0.018). The amino acid sequences in the regions which were studied did not change after 4 weeks of treatment. It is concluded that the presence of specific amino acids in position 2252 of PKRBD and position 2364 of V3 might be associated with clinical response to IFN. J. Med. Virol. 83:1332–1337, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis C virus (HCV) infection is one of the most frequent causes of chronic liver disease in many parts of the world, including Iran [Poynard et al., 2003; Merat et al., 2010]. Combination of pegylated interferon (IFN) and ribavirin (RBV) is the current standard treatment for chronic HCV infection. However, this treatment is not always successful, especially in patients infected with HCV genotype 1 [Manns et al., 2001]. Different host and viral factors have been shown to affect the response to treatment. Important viral factors include genotype and genetic heterogeneity of the virus [Poynard et al., 2003; Layden-Almer et al., 2005; Wohnsland et al., 2007]. A 12-amino acid sequence within the HCV E2 glycoprotein, termed the PKR/eIF2 phosphorylation homology domain (pePHD) may interact with the PKR and block its kinase activity which subsequently impairs the inhibition of protein synthesis [Cochrane et al., 2000; Hofmann et al., 2005]. It is hypothesized that mutations in the pePHD are related to response to IFN therapy [Gupta et al., 2006].
Positive correlation between the number of mutations in the IFN sensitivity-determining region (ISDR) of NS5A, and response to treatment has been reported especially from Japan. However, the results of some of the other studies from different parts of the world are controversial [Zeuzem et al., 1997; Chung et al., 1999; Berg et al., 2000; Jardim et al., 2009; Jenke et al., 2009].
Within the NS5A region of HCV, the PKRBD—a 66 amino acid sequence—is reported to influence the outcome of IFN based therapy through interaction with PKR [Sarrazin et al., 2000; Chisari, 2005].
Genetic heterogeneity in another region of the NS5A known as V3 might also affect the host response to treatment [Sarrazin et al., 2002].
In this study, the correlation between mutations in the pePHD, ISDR, PKRBD, and V3, and response to treatment in HCV 1a is investigated in isolates from Iranian patients. Mutations in these regions after 4 weeks of treatment were also studied.
METHODS
Study Population
Plasma samples were collected from 19 patients infected with HCV 1a who were referred to a University-based tertiary referral center affiliated to Tehran University of Medical Sciences. Samples were taken immediately before treatment and 4 weeks later.
Treatment Regimen
Patients received standard treatment with pegylated IFN-α2a and ribavirin for 48 weeks [Jabbari et al., 2010]. Sustained viral response was defined as undetectable HCV RNA in plasma by a sensitive method (lower limit of detection: 50 IU/ml) 24 weeks after the end of treatment [Shahzamani et al., 2010].
Amplification and Sequencing
HCV RNA was extracted and cDNA synthesized following standard procedure and nested PCR was used to amplify products for sequencing [Berg et al., 2000]. An alignment of 80 HCV 1a GenBank isolates was performed in order to identify suitable genotype 1a-specific segments. The primers were designed specifically to amplify the E2 and NS5A regions (Table I).
| Primer | Direction | Sequence | Position |
|---|---|---|---|
| E2 | |||
| P1 | F | 5′-ACTGATTGTTTCCGCAAGCATC-3′ | 2087–2109 |
| P2 | R | 5′-AGAATCCAAAGGGGTCCGAAG-3′ | 3025–3046 |
| P3 | F | 5′-TTCAAAGTCAGGATGTACGTG-3′ | 2219–2240 |
| P4 | R | 5′-TCTGGTCAGAAAATACTGAAGC-3′ | 2875–2897 |
| NS5A | |||
| P5 | F | 5′-CCCGAATTTTTCACAGAATTGG-3′ | 6717–6739 |
| P6 | R | 5′-TCCAGGAATAAGACATTGAGCAG-3′ | 7619–7642 |
| P7 | F | 5′-CTGATCCCTCCCATATAACAGC-3′ | 6889–6911 |
| P8 | R | 5′-AAGACATTGAGCAGCACACG-3′ | 7613–7633 |
- F, forward; R, reverse.
- * All primers were designed based on HCV 1a Ref. Seq. (NC_004102).
Appropriate dilution of first round products was used as a template for the second round of PCR. Direct sequencing was performed on the nested PCR products (SEQLAB, Göttingen, Germany).
All sequenced data were aligned and compared to the prototype sequence, HCV1a (GenBank accession No. M62321), using MEGA4 software [Tamura et al., 2007]. A consensus sequence was determined for each of the studied regions and individual sequences were compared against this sequence. The phylogenetic tree of the NS5A sequence obtained from HCV isolates was constructed by the Clustal-W method.
Statistical Analysis
Statistical analysis was performed using the Chi-square and Fisher's exact test. Analysis of continuous variables was performed using Student's t-test. A P value of less than 0.05 was considered significant. All analysis was performed using SPSS version 16.0 (SPSS, Inc., Chicago, IL).
Ethics
Written consent was obtained from all patients. Patient information was kept confidential and blinded from the researchers. The study protocol was approved by the institutional review board and the ethics committee of the Digestive Disease Research Center of Tehran University of Medical Sciences.
RESULTS
Some clinical features of patients are listed in Table II. There was no correlation between response to treatment and age, sex, viral load, and liver enzyme levels.
| Responders | Nonresponders | Total | |
|---|---|---|---|
| Number of subjects | 13 | 6 | 19 |
| Male/female | 11/2 | 6/0 | 17/2 |
| Age (years, mean ± SD) | 37.4 ± 11.6 | 46.2 ± 5.9 | 40.2 ± 10.8 |
| AST (IU/l, mean ± SD) | 46.6 ± 16.5 | 43.7 ± 18.4 | 45.7 ± 16.7 |
| ALT (IU/l, mean ± SD) | 92.9 ± 47.3 | 61.7 ± 16.9 | 83.1 ± 42.4 |
| Viral load (IU/ml, mean (range)) | 3,420,000 (12,570–18,600,000) | 5,540,000 (1,381,000–13,670,000) | 4,090,000 (12,570–18,600,000) |
- ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The amino acid sequence of the pePHD (codons 659–670) was compared with the consensus sequence of the 17 isolates and HCV 1a prototype (M62321). The alignment revealed that the pePHD is conserved in HCV 1a isolates.
Nine of the 13 patients who responded to treatment and five of the six nonresponders had no amino acid changes in the ISDR as compared to the consensus sequence (2209–2248). There was no significant difference between responders and nonresponders in number of mutations.
In the PKRBD region (2209–2274), the amino acid sequence of four responders and five nonresponders was identical to the consensus sequence (Fig. 1). The PKRBD sequence analysis showed more disagreement between responders and nonresponders than the ISDR sequence alone and even one of the nonresponders had three mutations within the PKRBD. This difference in number of mutations was not significant. However, at position 2252, seven out of 13 patients with a sustained response (53.8%) had replacement of isoleucine by valine compared to the consensus sequence whereas none of the patients without a sustained response had this mutation (P = 0.044).
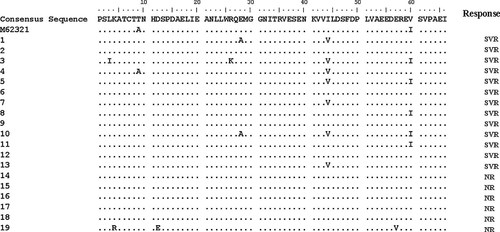
Amino acid sequence of the PKRBD region (2209–2274) aligned to the consensus sequence and prototype genotype 1a (M62321) (SVR, sustained viral response; NR, nonresponder).
The V3 region (2356–2379), located in the carboxyl terminus part of the NS5A, showed more variation especially in responders. All responders had amino acid substitutions in this region but only in two of the six nonresponders the sequence differed from the consensus (P < 0.001). The replacement of asparagine at position 2364 by aspartic acid or serine was observed in eight responders and none of the nonresponders (P = 0.018, Fig. 2).

Amino acid sequence of the V3 region aligned to the consensus sequence and prototype genotype 1a (M62321) (SVR, sustained viral response; NR, nonresponder).
In none of the regions studied did the pre-treatment sequence differ from the sequence obtained at week 4 of treatment.
Phylogenetic analysis of isolates showed no clustering among sequences (Fig. 3). Strains of five nonresponders (numbers 15–19) were located relatively close together but no uniform cluster was observed.
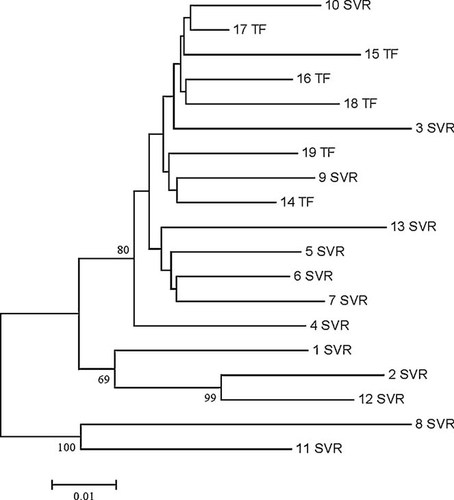
Phylogenetic tree of baseline NS5A sequences from 19 genotype 1a isolates (SVR, sustained viral response; NR, nonresponder).
DISCUSSION
Many researchers have studied viral and host factors that might affect the outcome of treatment [Yen et al., 2008]. Host factors, including innate immunity and genetic variations in IL28, have been shown to have an important role in control of HCV infection [Rauch et al., 2010]. One of the most important viral factors is the HCV genotype, but other viral factors are also involved. Analysis of some regions of the HCV genome has revealed differences in amino acid sequences between treatment responders and nonresponders. [Hofmann et al., 2005].
In the present study, it was observed that the amino acid sequence of the pePHD does not affect treatment response. This has also been reported by others [Polyak et al., 2000; Munoz de Rueda et al., 2008].
No consistent relationship has been observed between mutations in the NS5A region and response to treatment. Contradictory results from Japanese studies and those from other parts of the world indicate the importance of geographical and genetic factors [Pascu et al., 2004].
Enomoto et al. [1995] reported a significant relationship between IFN responsiveness and the amino acid sequence of the ISDR. They observed particularly that the amino acid in position 2218 had a critical effect on response to treatment. IFN-resistant HCV had histidine in this position whereas IFN-sensitive HCV did not. Similar reports concerning the effect of both number and type of amino acid substitutions and response to IFN therapy have been published [Watanabe et al., 2001]. However, conflicting results were observed by many European and American studies. These studies found no significant difference in the number and type of mutations in the NS5A region between responders and nonresponders [Kmieciak et al., 2006; Brillet et al., 2007].
In a study from Japan, HCV genotype 1b was classified into three subtypes: Japanese specific J-type, Worldwide W-type, and a third NJ-type that is not found in Japan. As the ISDR mutation rate, as well as response to treatment, was higher in the J-type HCV than the W-type, the authors suggested that geographic factors predicted the outcome of treatment through variations in the ISDR sequence [Murayama et al., 2007].
Variations in amino acids at positions 2217 and 2218 of the ISDR have been reported previously. These positions were suggested as critical points in the phosphorylation of the NS5A and its interaction with cellular factors [Nousbaum et al., 2000; Schiappa et al., 2002].
In the present study, the ISDR and PKRBD—a 26 amino acid sequence downstream to ISDR—were analyzed in relation to treatment response. Although a difference was observed in the number of mutations between patients with and without a sustained viral response, this did not reach statistical significance. However, it was observed that the change from isoleucin to valine at position 2252, as compared to the consensus sequence, was positively correlated with a sustained viral response. Similarly, the variability in the V3 region was associated with a sustained viral response, as already reported by others [Murphy et al., 2002; Dal Pero et al., 2007; Jardim et al., 2009]. A significantly higher number of mutations was observed in the current study in responders. Geographic and ethnic factors might also be involved. In a study by Layden-Almer et al. [2005] on African–Americans and white patients, significantly more mutations were observed in the V3 region of HCV isolated from white patients. These patients also responded better to treatment. In accord with the present study, Layden-Almer et al. observed that the presence of asparagine at position 2364 within the V3 region was associated with poor treatment response.
In the present study, the pretreatment sequence of the pePHD, ISDR, PKRBD, and V3 regions was compared with isolates taken from the same patients 4 weeks after start of treatment. The fact that no mutations were observed suggests that positive selection of treatment-resistant variants does not occur during IFN therapy, at least during the first 4 weeks. This has also been reported in other studies [Schiappa et al., 2002].
It is concluded that the presence of specific amino acids in position 2252 of the PKRBD region and position 2364 of V3 might be associated with clinical response to IFN.
Acknowledgements
The authors are indebted to Elham Fakhar-zadeh and Hedyeh Zamini for their help in sample collection and clinical follow-up. We also express our gratitude to Hooman Khademi for helping us with the statistical analysis and Sadaf Ghajarieh Sepanlou for reviewing the manuscript.




