Comparative analysis of disease severity between genotypes IA and IIIA of hepatitis A virus†
Min Ja Kim and Byung Chul Chun contributed equally to this work.
Abstract
Although hepatitis A is a major health problem worldwide, it has not yet been clarified whether or not viral factors affect the clinical characteristics. This study aimed to investigate if a genotype of hepatitis A virus (HAV) affects disease severity among adolescent and adult populations. Clinical data and specimens were collected from patients ≥16-years-of-age with acute hepatitis A at two university hospitals in Korea during the two study periods: 1998 and 1999 (n = 45), and 2009 (n = 66). Nucleotide sequencing of the complete VP1 region of the HAV isolates was performed for phylogenetic analysis and genotyping. Clinical parameters related to disease severity were compared by HAV genotype to determine its clinical relevance. Of the 87 patients, 47 were male and the mean age was 29.8 ± 8.1 years. The genotype IIIA (93.0%, 53/57) was predominant in the year 2009, whereas IA (93.3%, 28/30) was the major genotype in 1998 and 1999. When comparing disease severity between the two HAV genotypes, the patients with genotype IIIA were older and had higher alanine aminotransferase (ALT) levels, prolonged prothrombin times and lower serum albumin levels. In a multivariate logistic regression model, higher ALT levels ≥ 1,000 IU/L (odds ratio [OR] 11.7, 95% confidence interval [CI] 2.5–54.0) and longer hospitalization (OR 22.49, 95%CI 4.6–132.5) were associated independently with genotype IIIA. In conclusion, this study indicates that HAV genotype might be one of the viral factors responsible for the disease severity of hepatitis A. J. Med. Virol. 83:1308–1314, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis A virus (HAV) infection is a significant public health problem throughout the world. In the Republic of Korea, the seroprevalence of anti-HAV antibodies among teens and those aged 20–29 years has declined rapidly to <20% in 2006 [Sohn et al., 2000; Lee et al., 2008]. As a result of the epidemiological shift, clinically overt HAV infection has increased among adolescents and adults since 2006 [Korea Centers for Disease Control and Prevention, 2008; Kim and Lee, 2010], and patients with severe presentations and the accompanying complications have been reported increasingly [Shin and Kim, 2009; Jung et al., 2010; Kim et al., 2010]. HAV infection has been notified as an officially designated communicable disease in the Republic of Korea since 2009.
HAV is a non-enveloped RNA virus with a single-stranded 7.5 kb genome [Najarian et al., 1985]. The viral capsid is composed of three exposed polypeptides (VP1, VP2, and VP3) and a putative VP4, with a high degree of antigenic and genetic conservation, as reflected by the existence of a single serotype [Gauss-Muller et al., 1986]. A HAV genotype is defined as a group of viruses with a >85% nucleotide sequence identity, and is further classified into subgenotypes with a sequence variability of <7.5% [Robertson et al., 1992]. Based on sequences of the VP1/2A genomic region, different HAV strains have been classified into seven genotypes (I–VII) which are distinguishable by 15–25% sequence diversity. Four of the genotypes (I, II, III, and VII) are of human origin [Robertson et al., 1992; Ching et al., 2002]. On the other hand, a study of South American HAV strains reveals that the VP1 amino terminus, as an immunodominant site, contains more informative variable positions than the VP1/2A junction [Costa-Mattioli et al., 2001].
Several studies on HAV genotypes in the Republic of Korea have shown a distinct changing pattern in circulating HAV genotypes over the past 10 years. Until early 2000, almost all isolates tested had been identified as genotype IA [Byun et al., 2001]. However, studies conducted from 2004 to 2008 reported the co-circulation of the two prevalent genotypes IA and IIIA [Yoo et al., 2008; Yun et al., 2008; Park et al., 2009; Song et al., 2009]. More recent reports showed that genotype IIIA was the predominant serotype since 2008 [Lee et al., 2009; Yoon et al., 2009]. An increase in the number of adult cases of clinically overt hepatitis A, together with increasing proportion of genotype IIIA in the Republic of Korea, gave us opportunities to investigate the relationship between the HAV genotype and disease severity.
It is considered that the disease severity of HAV infection might be influenced by the patient age and any underlying chronic liver disease [Vento et al., 1998]. Besides the host factors, viral factors such as viral load and genomic differences have been considered to be factors related to the varying degrees of disease severity [Rezende et al., 2003; Yokosuka, 2005]. However, correlation between the HAV genome and the clinical status of HAV infection has not yet been established.
The aim of this study was to compare the clinical features between HAV genotypes IA and IIIA, and to determine whether or not the HAV genotype affected disease severity in adolescent and adult populations.
METHODS
Study Population and Data Collection
The study population included patients from two university hospitals who were older than 16-years-of-age and who had symptoms compatible with acute hepatitis A, positive anti-HAV IgM and available clinical specimens for HAV genotyping. To be able to compare the two genotypes IA and IIIA, clinical samples collected in 1998 and 1999 (45 serum specimens) and in 2009 (66 stool specimens) were included. Demographic and clinical data obtained included white blood cell (WBC) and platelet counts, blood urea nitrogen (BUN), creatinine (Cr), aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, gamma-glutamyl transpeptidase (GGT), international normalized ratio (INR) of prothrombin time (PT), albumin, and C-reactive protein (CRP). Clinical outcome and complications such as acute kidney injury, hepatic encephalopathy, or myocarditis were also evaluated.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Stool specimens were diluted to a ratio of 1:10 in phosphate-buffered saline, mixed, and centrifuged (4,000 rpm, 10 min, 4°C). Viral RNA was extracted from either a 200 µl serum sample or a fecal suspension using the QIAamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany), and cDNA was generated with an AccuPower® RT/PCR PreMix system (Bioneer, Alameda, CA).
To amplify the complete 900-bp sequence of the VP1 genomic region, three pairs of the VP1-specific primers were used as previously described [Yoon et al., 2010]. The amplified products were confirmed on 2% agarose gel electrophoresis. Single step RT-PCR was carried out initially using either a set of the VP1-1N-F and VP1-2-R primers, or alternatively, a pair of the VP1-A-F and VP1-A-R. When the single step RT-PCR failed to amplify the PCR products, the nested PCR was followed using a set of the VP1-AN-F and VP1-AN-R. All amplification reactions were carried out at 42°C for 60 min, 94°C for 5 min, 40 cycles of 94°C for 1 min, 50°C for 1 min and 1 min at 72°C and elongation at 72°C for 7 min.
Nucleotide Sequencing and Genotyping
RT-PCR products were purified using the PCR quick-spin PCR product purification kit (Intron Biotechnology, Seoul, Korea). The analysis of nucleotide sequences was carried out by using the BigDye® Terminator v 3.1 Kit (Applied Biosystems, Foster City, CA) and the ABI 3730 × l Genetic Analyzer of Capillary electrophoresis system (Applied Biosystems). The genotype was determined through phylogenetic analysis of the complete VP1 sequence. The VP1 sequences of the HAV isolates were aligned with those of the reference HAV genotypes available in the GenBank by using Clustal W2 software (http://www. Wbi.ac.uk/clustalw). Phylogenetic trees were constructed using the MEGA software by the neighbor-joining method from a Kimura two-parameter distance matrix. Their reliability was assessed by bootstrap resampling (1,000 pseudoreplicas).
Nucleotide Sequence Accession Numbers
The complete sequences of the VP1 region from the HAV isolates in this study were registered in the GenBank under the accession numbers GU908224–GU908289. The reference strains (accession number) for the specific genotype included, respectively, NOR-21 (AJ299464, Norway), KNIH-ISJ (EU849137, Korea), HA-JNG04-90 (AB279732, Japan) and PN-IND (EU011791, India) for genotype IIIA, KNIH-KEO (EU849135, Korea), Par05.02/B (DQ504426, Spain) and KNIH-KSJ (EU849136, Korea) for genotype IA, HM-175 (M14707, Australia), HAF-203 (AF268396, Brazil) and MBB (M20273, German) for genotype IB, SLF88 (AY644670, USA) for genotype IIB, and HA-JNG06-90F (AB258387, Japan) and HAJ85-1 (AB279735, Japan) for genotype IIIB.
Statistical Analyses
Descriptive statistics were used to describe the study population and the proportion of HAV genotypes. Differences in clinical and laboratory parameters associated with disease severity by the HAV genotypes were assessed using Chi-square tests for categorical variables and Student's t-test for continuous variables. Fisher's exact test was used where an expected cell value was <5. A multivariable logistic regression model was used to determine adjusted odds ratio (OR) and a 95% confidence limit for each parameter, adjusted for age, sex, and the duration between the onset of illness and hospitalization. All statistical analyses were conducted using SPSS version 14.0 (SPSS, Chicago, IL).
RESULTS
General Characteristics of Patients With Hepatitis A
Of 111 patients examined, 87 patients who were determined to have been infected by the HAV genotype were included in the present comparative analyses. Forty-seven patients were male. The mean age was 29.8 ± 8.1 years (range, 16–49 years). The distribution of age, year of diagnosis and genotype are shown in Table I. Duration of the onset of illness before admission was 5.1 ± 2.8 days (range, 1–15 days). The duration of hospitalization was 15.2 ± 9.4 days (range, 0–42 days). One patient was co-infected with hepatitis B virus and one with hepatitis C virus.
| Characteristics | Classification | n | % |
|---|---|---|---|
| Sex | Male | 47 | 54.0 |
| Age | 15–19 | 8 | 9.2 |
| 20–29 | 35 | 40.2 | |
| 30–39 | 34 | 39.1 | |
| 40–49 | 10 | 11.5 | |
| Year of diagnosis | 1998–1999 | 30 | 34.5 |
| 2009 | 57 | 65.5 | |
| Genotype | IA | 32 | 36.8 |
| IIIA | 55 | 63.2 |
HAV Genotyping and Sequence Analyses
HAV genotype was identified in 30 patients (66.7%) during 1998 and 1999, and 57 patients (86.3%) during 2009. Construction of a phylogenetic tree included 65 different HAV strains obtained from this study and 13 reference strains retrieved from Genbank (Fig. 1). The complete VP1 sequence was identical in 8 of 32 genotype IA strains, whereas the sequence was identical in 14 of 55 genotype IIIA isolates. Proportions of genotypes IA and IIIA in 1998 and 1999 were 93.3% (n = 28) and 6.7% (n = 2) respectively. In contrast, in the year 2009 genotypes IA and IIIA accounted for 7.0% (n = 4) and 93.0% (n = 53), respectively. This result indicates an apparent increase in the proportion of genotype IIIA in 2009 (P < 0.001).
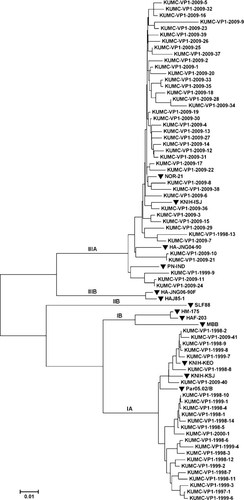
The phylogenetic tree constructed for the nucleotide sequences of VP1 genomic region of hepatitis A virus. The tree includes 65 strains isolated from this study (GU908224–GU908242, GU908244–GU908289) and the reference strains {IIIA: NOR-21 (AJ299464, Norway), KNIH-ISJ (EU849137, Korea), HA-JNG04-90 (AB279732, Japan), PN-IND(EU011791, India); IA: KNIH-KEO (EU849135, Korea), Par05.02/B (DQ504426, Spain), KNIH-KSJ (EU849136, Korea); IB: HM-175(M14707, Australia), HAF-203 (AF268396, Brazil), MBB (M20273, German); IIB: SLF88 (AY644670, USA); IIIB: HA-JNG06-90F (AB258387, Japan), HAJ85-1 (AB279735, Japan)}. (▾) Reference strains.
The genotype IA isolates from the two study periods showed 94–100% sequence homology. They were also related closely to the Par05.02/B (Spain), KNIH-KEO (Korea), and KNIH-KSJ (Korea) as the reference strains, with 97–99% identity. Similarly, the analyzed genotype IIIA isolates displayed 94–100% homology to each other, and were related closely to NOR-21 (Norway), KNIH-ISJ (Korea), HA-JNG04-90 (Japan) and PN-IND (India), with 94–99% identity.
Comparative Analysis of Disease Severity Between Genotype IIIA and Genotype IA
Compared to those with genotype IA, cases with genotype IIIA were older and had higher ALT, AST, GGT, bilirubin and Cr levels, prolonged PT, and lower serum albumin and platelet levels in univariate analysis (Table II). But, there was no difference in duration between the onset of illness and hospitalization.
| Variables | Genotype | Pa | |
|---|---|---|---|
| IIIA (n = 55) | IA (n = 32) | ||
| Age, years | 33.6 ± 6.7 | 23.3 ± 5.9 | <0.001 |
| Male, n (%) | 32 (58.2) | 16 (48.5) | 0.253 |
| Diagnosis in 2009, n (%) | 53 (96.4) | 4 (12.5) | <0.001 |
| HBV, n (%) | 1 (1.8) | 0 (0.0) | 0.632 |
| HCV, n (%) | 0 (0.0) | 1 (3.1) | 0.368 |
| Interval between the onset and hospitalization, days | 4.7 ± 2.7 | 5.9 ± 2.9 | 0.053 |
| Duration of hospitalization, days | 19.8 ± 7.8 | 7.5 ± 6.3 | <0.001 |
| BUN, mg/dl | 15.2 ± 21.5 | 9.3 ± 3.6 | 0.057 |
| Cr, mg/dl | 1.9 ± 3.3 | 0.8 ± 0.2 | 0.024 |
| WBC, ×109/L | 4.8 ± 2.1 | 4.6 ± 1.5 | 0.616 |
| Platelets, ×109/L | 158.6 ± 69.1 | 204.4 ± 86.5 | 0.013 |
| AST, IU/L | 2,836.4 ± 2,617.0 | 1,492.0 ± 1,338.3 | 0.002 |
| ALT, IU/L | 3,102.7 ± 1,891.8 | 1,089.0 ± 1,637.4 | <0.001 |
| Bilirubin, mg/dl | 8.2 ± 6.5 | 5.0 ± 3.2 | 0.002 |
| PT, INR | 1.3 ± 0.4 | 1.1 ± 0.1 | <0.001 |
| GGT, IU/L | 376.6 ± 236.0 | 260.8 ± 154.1 | 0.030 |
| CRP, mg/L | 22.9 ± 22.8 | 12.8 ± 19.8 | 0.099 |
| Albumin, g/dl | 3.6 ± 0.4 | 4.1 ± 0.4 | <0.001 |
- HBV, hepatitis B virus infection; HCV, hepatitis C virus infection; BUN, blood urea nitrogen; Cr, creatinine; WBC, white blood cell; AST, aspirate aminotransferase; ALT, alanine aminotransferase; PT, prothrombin time; INR, international normalized ratio; GGT, gamma-glutamyl transferase; CRP, C-reactive protein.
- a Adjusted by age and sex.
In multivariate analysis, higher ALT level (≥1,000 IU/L; OR 11.7, 95% confidence interval [CI] 2.5–54.0) and longer hospitalization (OR 22.49, 95%CI 4.6–132.5) were the independent severity indices associated with genotype IIIA (Table III).
| Severity indices | Genotype, n (%) | cOR (95%CI) | aORa (95%CI) | |
|---|---|---|---|---|
| IIIA | IA | |||
| ALT ≥1,000 IU/L | 49 (89.1) | 9 (28.1) | 20.9 (6.7, 65.6) | 11.7 (2.5, 54.0) |
| ALT ≥2,000 IU/L | 36 (65.5) | 6 (18.8) | 8.2 (2.9, 23.4) | 4.5 (1.2, 17.6) |
| Hospitalization ≥14 days | 42 (80.8) | 4 (12.9) | 28.4 (8.1, 99.6) | 24.7 (4.6, 132.5) |
| Platelets <150 × 109/L | 31 (56.4) | 11 (33.3) | 2.7 (1.2, 6.2) | 2.2 (0.7, 7.0) |
| Bilirubin ≥5 mg/dl | 38 (69.1) | 12 (37.5) | 3.7 (1.5, 9.3) | 3.4 (0.9, 13.0) |
| GGT ≥300 IU/L | 21 (70.0) | 9 (31.0) | 5.2 (1.7, 15.7) | 3.5 (0.8, 15.9) |
| WBC <4.5 × 109/L | 32 (58.2) | 17 (53.1) | 1.2 (0.5, 3.0) | 0.8 (0.2, 2.7) |
| CRP ≥20 mg/L | 20 (38.5) | 2 (11.1) | 5.0 (1.0, 24.1) | 1.5 (0.2, 14.2) |
| PT <40% | 6 (10.9) | 1 (3.4) | 3.4 (0.4, 30.0) | 4.2 (0.2, 82.8) |
| Albumin <3.5 g/dl | 16 (30.8) | 3 (10.0) | 4.0 (1.1, 15.1) | 3.4 (0.6, 20.0) |
| Acute kidney injury, n (%) | 6 (11.1) | 0 (0.0) | — | — |
| Extrahepatic complication, n (%) | 6 (11.1) | 1 (3.6) | 3.1 (0.3, 27.4) | 0.7 (0.1, 9.3) |
| Deaths, n (%) | 2 (3.6) | 0 (0.0) | — | — |
- ALT, alanine aminotransferase; GGT, gamma-glutamyl transferase; WBC, white blood cell; CRP, C-reactive protein; PT, prothrombin time.
- a OR, odds ratio; adjusted by age, sex and interval between the onset of illness and hospitalization.
Of the 57 patients in 2009, two died; one from bacterial sepsis and another from disseminated intravascular coagulation. Two additional cases presented with hepatic encephalopathy (n = 1) and myocarditis (n = 1). Six (9.1%) were complicated with acute renal injury. In contrast, no deaths or cases with remarkable complications occurred in 1998 and 1999.
DISCUSSION
Whether or not the HAV genotype has an effect on the disease severity of hepatitis A in adolescent and adult populations was investigated. Clinical parameters of severity between two patient groups infected with HAV genotypes IIIA and IA strains were compared. A higher ALT level (≥1,000 IU/L) and longer hospitalization were associated significantly with genotype IIIA. In addition, this genotype was the predominant circulating genotype among the HAV isolates in 2009, supporting the findings on the increase of HAV genotype IIIA isolates in the Republic of Korea in recent years [Lee et al., 2009; Yoon et al., 2009].
With the advance in technology of molecular virology, several studies have investigated whether or not viral factors such as nucleotide substitutions, viral load and genotype might affect the clinical characteristics of hepatitis A. Fujiwara et al. [2002, 2003, 2007, 2009] carried out studies on the substitution of HAV sequences from patients with hepatitis A in various degree of severity. They suggested that differences in HAV genetic variations of 5′ non-coding regions 2B and 2C between fulminant and self-limited hepatitis A might cooperatively influence viral replication and virulence. Mackiewicz et al. [2010] reported the association of mutations in the internal ribosome entry site of HAV with enhanced in vitro replication, despite lack of relevance to a clinical phenotype. Several studies reported that the viral load might be closely correlated with liver damage and disease severity in mild cases of hepatitis, but not in severe cases [Rezende et al., 2003; Sainokami et al., 2005; Fujiwara et al., 2009]. In addition, Ajmera et al. [2010] studied host and viral factors in 29 adult patients with HAV-associated acute liver failure. No significant differences in the rates of single nucleotide or transcribed amino acid residue substitutions in the complete HAV genome were evident between patients with HAV-associated acute liver failure and those with uncomplicated acute hepatitis. The study also determined that more frequent viral clearance, as defined by PCR, was associated with poor outcome in acute liver failure as well as the finding of familial cases, suggesting a possible host genetic predisposition. Furthermore, Fujiwara et al. [2011] reported that patients having a severe infection such as fulminant hepatitis and severe acute hepatitis had a higher initial viral load, compared to patients with less severe infection (P < 0.001), and suggested that a resultant excessive host immune response could reduce the viral load rapidly as a result of the destruction of large numbers of HAV-infected hepatocytes.
There is little available information on the clinical relevance of HAV genotype. Rezende et al. [2003] reported that genotype IA was detected more frequently in cases with a milder course of hepatitis A compared to fulminant hepatitis A. Fujiwara et al. [2003] failed to show any role of the HAV genotype in patients with varying severities of hepatitis A, because only one or two numbers of different genotypes (IB or IIIA) were included. Hussain et al. [2005] reported that HAV genotypes IA and IIIA were equally prevalent in northern India, possibly reflecting an epidemiological shift or geographical variation. The study also demonstrated that clinical severity, based on the mean liver-function profile, was correlated significantly with positivity for anti-HAV IgM but did not differentiate the severity at the subgenotype level. In particular, there have been no studies comparing disease severity between HAV genotypes IA and IIIA.
In this study, multivariate analysis showed that patients infected by the hepatitis A virus genotype IIIA displayed a significantly higher risk of ALT ≥1,000 IU/L and were more apt to experience longer hospitalization than those infected by genotype IA, with aORs of 11.7 and 24.7, respectively. However, the 95%CI of both ratios were considerably wide due to the small sample size. The ORs of prolonged PT (<40%), acute kidney injury, hyperbilirubinemia (≥5 mg/dl), extra-hepatic complications and death were not significant, due to the small event number in genotype IA patients. Other clinical parameters such as platelets <150 × 109/L, bilirubin ≥5 mg/dl, GGT ≥300 IU/L, CRP ≥20 mg/L, and albumin <3.5 g/dl had significant cORs exceeding 1.0. However, the aORs adjusted by age, sex and the duration between the onset of illness and hospitalization were not significant with wide CIs. These findings might result from the small sample size, the effect of the adjusted variables or both. Especially, patient age, as one adjusted variable, had a strong influence on OR in multivariate analysis. But, aORs of ALT ≥1,000 IU/L, and hospitalization ≥14 days were still significant after the adjustment. These results suggest that genotype IIIA might be more virulent than genotype IA.
This study has limitations primarily related to its descriptive nature. The small sample size and the small number of events made the CI wide. During the years of diagnosis (1998–1999 and 2009) for patient collection, there were no remarkable changes on health systems, hospitalization policy and treatment methods of acute hepatitis in the Republic of Korea. Therefore, it is unlikely that this 10-year interval might alter the statistical significance of the clinical differences between genotypes IIIA and IA. Further studies on how genotype IIIA affects the disease severity of hepatitis A remain to be done, including virulence of different genotypes in cell line infections [Emerson et al., 2002].
Of the several HAV genotypes, genotype I is the most common worldwide. Genotype IA is predominant in North America, Europe, China, Japan, America, Russia, and Thailand [Costa-Mattioli et al., 2003], whereas genotype IIIA has a history of continuous circulation in India, Sri Lanka, Nepal, and Malaysia [Khanna et al., 1992]. HAV strains representing one genotype can circulate over a long period of time without any great changes in the nucleotide sequences [Arauz-Ruiz et al., 2001; Arankalle et al., 2006]. This study suggests that the major circulating genotype has been switched to genotype IIIA from the genotype IA in the Republic of Korea in recent years, probably due to the spread of the pre-existing genotype IIIA and the increasing numbers of the susceptible populations in the community.
In conclusion, the current study indicates that HAV genotype IIIA, compared to genotype IA, is associated significantly with higher ALT levels and longer hospitalization in adolescent and adult patients, suggesting that the HAV genotype might be, at least in part, responsible for disease severity. To our knowledge, this is the first study to establish HAV genotype as one of the potential viral factors responsible for the severity of HAV infection.




