Conservation and mutation of viral interleukin-10 gene in gastric carcinomas and nasopharyngeal carcinomas†
Institution where the work was performed: Department of Medical Microbiology, Qingdao University Medical College, Qingdao, China
Abstract
The Epstein–Barr virus (EBV) BamHI-C fragment rightward reading frame 1 (BCRF1)-coded viral interleukin-10 (vIL-10), exhibits high homology with human interleukin-10 (hIL-10) gene. The protein product vIL-10, which shares some functional properties with hIL-10, primarily mediates immunosuppressive functions. To characterize the variations of the vIL-10 gene and to explore the association between vIL-10 gene variations and EBV associated diseases, the vIL-10 gene was analyzed (using direct sequencing) in 41 cases of EBV-associated gastric carcinoma (EBVaGC), 83 nasopharyngeal carcinoma biopsies, and 40 throat washing samples from healthy donors in Northern China. One silent mutation (c9980a) was observed in the majority of EBVaGC, nasopharyngeal carcinoma and throat washing samples (134/164, 81.7%). Two consensus mutations (V6M and G23S) were identified in the signal peptide region in some nasopharyngeal carcinoma and throat washing isolates. These results indicate that the pattern B95-8 (identical sequence to B95-8) is the dominant type among the EBV isolates from Northern China, while the pattern SPM (mutation in the signal peptide present only in nasopharyngeal carcinoma and throat washing isolates) seems more relevant with the EBV-positive nasopharyngeal and laryngopharyngeal epithelial cells. The conservation of vIL-10, with a few variations, suggests the critical role of the vIL-10 gene for EBV in gaining an advantage over the host's immune system. J. Med. Virol. 83:644–650, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Epstein–Barr virus (EBV) infects approximately 95% of the adults world wide. After initial exposure to EBV, most individuals develop asymptomatic infections and remain to be lifelong carriers of the virus. However, EBV may cause infectious mononucleosis (IM) and is closely associated with several malignancies including Burkitt's lymphoma (BL), Hodgkin lymphoma (HD), nasopharyngeal carcinoma, and gastric carcinoma [Niedobitek, 2000; Thompson and Kurzrock, 2004; Wang et al., 2005; Bornkamm, 2009].
The EBV gene BamHI-C fragment rightward reading frame 1 (BCRF1) encodes viral interleukin-10 (vIL-10), which has a strong homology to the murine and human interleukin-10 (mIL-10 and hIL-10) genes [Moore et al., 1990; Vieira et al., 1991]. This relationship is most pronounced in the mature protein-coding sequences, where hIL-10 and BCRF1 are 84% identical, and does not appear in the signal sequence or flanking untranslated regions [Moore et al., 1991]. The vIL-10 gene is expressed late in the lytic phase of virus replication and also early in EBV infection of human B cells [Hudson et al., 1985; Miyazaki et al., 1993; Stewart et al., 1994; Touitou et al., 1996; Sairenji et al., 1998]. The vIL-10 product shares some functional properties with hIL-10, predominantly relating to its immunosuppressive function [Hsu et al., 1990; Vieira et al., 1991]. The vIL-10 inhibits cytokine synthesis (interleukin-2 and interferon-γ) by human peripheral blood mononuclear cells (PBMCs) [Fiorentino et al., 1989; Fiorentino et al., 1991; Taga and Tosato, 1991]. In the murine model, infected with a recombinant vaccinia virus expressing IL-10, the hIL-10 and vIL-10 may provide a selective advantage by blunting the early human natural killer cell and cytotoxic T-cell responses, so EBV can establish a well-contained latent infection in B lymphocytes [Kurilla et al., 1991]. In addition, vIL-10 strongly reduces antigen-specific human T-cell proliferation by diminishing the antigen-presenting capacity of monocytes via down-regulation of class II major histocompatibility complex (MHC) expression [de Waal Malefyt et al., 1991]. However, in contrast to hIL-10, vIL-10 is unable to costimulate thymocyte and mast cell proliferation and does not induce B-cell class II MHC expression [Go et al., 1990; MacNeil et al., 1990; Vieira et al., 1991]. The partial differences in functional properties between vIL-10 and hIL-10 might be the result of constant selective pressure on EBV to gain an advantage over the host's immune system.
In China, most studies on sequence variations of the EBV genome have focused on the populations in Southern China, the endemic area of nasopharyngeal carcinoma [Zhang et al., 2002; Chang et al., 2009; Li et al., 2009; Chen et al., 2010]. However, there is no reported polymorphism in the vIL-10 gene in China. To the best of our knowledge, only one study with a small number of cases indicated the conservation of vIL-10 gene in several EBV-positive cell lines, EBV-associated gastric carcinoma (EBVaGC) and throat washing isolates in Japan [Kanai et al., 2007]. There is no report regarding polymorphisms in vIL-10 gene on EBV-positive nasopharyngeal carcinoma samples. This study analyzed the nucleotide sequences of vIL-10 gene and determined the variations of this gene in EBVaGC, nasopharyngeal carcinoma and throat washing samples in Northern China, where nasopharyngeal carcinoma is not endemic.
MATERIALS AND METHODS
Sample Collection and DNA Extraction
This study was approved by the Medical Ethics Committee at the Medical College of Qingdao University, China. Informed consent was obtained from all subjects infected with EBV who participated in the study. Fresh or paraffin-embedded gastric carcinoma tissues and paraffin-embedded nasopharyngeal carcinoma tissues were collected from seven county hospitals of the Shandong Province in the Northern part of China. The EBV-associated gastric carcinoma and nasopharyngeal carcinoma samples were screened by in situ hybridization for EBV-encoded small RNA1 (EBER1), as described previously [Sugiura et al., 1996]. Throat washing samples were collected from healthy donors in the same geographical regions by gargling with 15 ml of phosphate buffered saline. PCR with a BamHI W-specific primer pair was used to screen the throat washing samples for the EBV genome [Ikuta et al., 2000].
DNA was extracted from fresh tumor tissues and throat washing samples by using the standard method with proteinase K digestion and phenol-chloroform purification. QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany) was used to extract DNA from paraffin-embedded tumor tissues.
PCR and DNA Sequencing
Nested-polymerase chain reaction (nested-PCR) was performed to determine the sequence variation in the vIL-10 gene (nucleotide sequence 9675-10187; Table I). The first PCR was carried out with BCRF1-1 and BCRF1-2 in a total volume of 25 µl containing 1× PCR reaction buffer, 100 ng of genomic DNA, 0.5 µM of each primer, 200 µM of each deoxyribonucleotide triphosphates, and 1 U Pfu Taq polymerase (TaKaRa Biotechnology, Kyoto, Japan). The cycling parameters were 94°C for 5 min; 35 cycles of 94°C for 30 sec, 53°C for 30 sec, 72°C for 1 min; and a final elongation step at 72°C for 10 min. The products of the first round of PCR were amplified in the second round of PCR with BCRF1-3 and BCRF1-4, except for annealing at 50°C. In each set of PCR, DNA from EBV-positive B95-8 cell lines and EBV-negative Ramos cell lines were used as positive and negative controls, respectively. In addition, several measurements were taken to prevent contamination during each experiment, such as frequently changing gloves and cleaning the equipment, using aerosol-resistant pipette tips for PCR, and performing different procedures in separate areas.
| Name | Sequence (5′–3′) | Analyzed region (B95.8 coordinates) |
|---|---|---|
| BCRF1-1 | CGCGGGAAATACGTCCTAC | 9517–9535 |
| BCRF1-2 | TACCTGGGCGGCAAGAAC | 10311–10294 |
| BCRF1-3 | TGTAAAAATAGAACGCCC | 9568–95866 |
| BCRF1-4 | GTAAATGACAGTCGAAAGG | 10238–10257 |
After amplification, the PCR products were purified using a gel extraction kit (Qiaex II; Qiagen, Hilden, German) and sequenced directly in both directions with the primers BCRF1-3 and BCRF1-4 by a Prism ready reaction Dyedeoxy terminator cycle sequencing kit (Applied Biosystems, Foster, CA). The PCR product of B95-8 cells was simultaneously sequenced to ensure the integrity of the run. The sequences from partial isolates, especially all unique or rare variants were confirmed by analyzing a second PCR product.
The sequence data were checked for homology using BLAST (National Center for Biotechnology Information: NCBI; http://www.ncbi.nlm.nih.gov) and were compared with the B95-8 prototype strain (GenBankacc.No.V01555). Alignments between sequences were analyzed using DNAStar software (Larsergene, version 7.0).
Statistical Analysis
The Fisher's exact test was performed to determine the difference of the EBV genotypes among the EBV-associated gastric carcinoma, nasopharyngeal carcinoma and healthy donors. Significance was set at P < 0.05. Statistical analyses were conducted using SPSS 15.0 statistical software (SPSS, Chicago, IL).
RESULTS
The vIL-10 gene (nucleotide sequence 9675–10187; amino acid sequence 1–170) was successfully amplified and sequenced in 164 EBV-positive samples, including 41 EBV-associated gastric carcinoma isolates, 83 nasopharyngeal carcinoma biopsies and 40 throat washing samples from healthy donors. Analysis by DNA direct sequencing showed the presence of a single vIL-10 gene sequence in all of 164 cases (100.0%).
The vIL-10 genes encode 170 amino acids including the signal peptide (aa 1–25) and mature protein (aa 26–170). Signal peptides typically consist three domains, a positively charged (n-) region, a hydrophobic core (h-) region, and a polar carboxy-terminal (c-) region [Perlman and Halvorson, 1983; von Heijne, 1985]. The vIL-10 mature protein consists of six helices (A–F) and five loops between the helices. Their receptor-binding sites are located at helices A and F and the loop between helix A and helix B (AB loop) [Zdanov et al., 1997; Yoon et al., 2005]. The mutations in the vIL-10 gene were mapped on the amino acid sequence of the vIL-10 protein (Figs. 1 and 2).
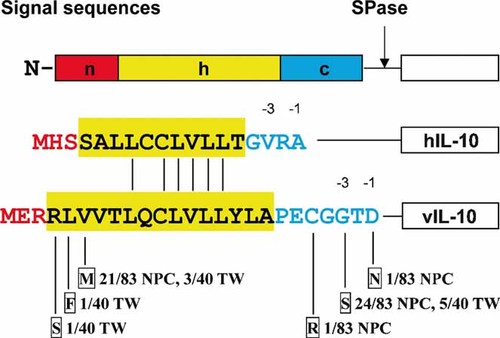
Amino acid mutations in signal peptide region of vIL-10 in wild isolates from EBV-positive nasopharyngeal carcinoma biopsies and throat washing samples from healthy donors. The tripartite structures of signal peptide are indicated by different colors: a central hydrophobic h-region (yellow), hydrophilic N- (red), and C-terminal (blue) flanking regions. Identical residues between hIL-10 and vIL-10 are shown by short vertical lines. The mutated amino acid residues to signal sequences of vIL-10 are indicated in boxes and the mutated numbers of the isolates were indicated in the proportions. Arrow: cleavage site for signal peptidase (SPase). NPC, means nasopharyngeal carcinoma; TW throat washing samples from healthy donors.
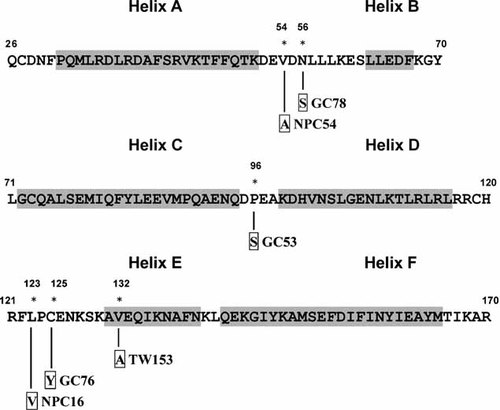
Amino acid mutations in mature protein of the vIL-10 in wild isolates from EBV-associated gastric carcinoma, nasopharyngeal carcinoma biopsies, and throat washing samples from healthy donors. The vIL-10 amino acid sequence of B95-8 is shown in capital letters. Helix structures from A to F are shown in gray quadrangles. Numbers indicate the amino acid positions, and the asterisks indicate the mutant amino acids. The mutated amino acid residues to the vIL-10 mature protein are indicated in boxes and the mutated isolates are indicated by the name of the samples. GC, EBV-associated gastric carcinoma; NPC, nasopharyngeal carcinoma; TW throat washing samples from healthy donors.
Conservation of Viral Interleukin-10 Gene
According to DNA Star MegAlign, the homology of the EBV-positive gastric carcinoma, nasopharyngeal carcinoma, and throat washing sequences exceeded 98.8% (EBV-associated gastric carcinoma isolates ≥99.6%, nasopharyngeal carcinoma biopsies ≥98.8% and throat washing samples ≥99.2%). Compared with the B95-8 prototype sequence, no sequence change was found in 29 specimens (17.7%), including 7 EBV-associated gastric carcinoma isolates (17.1%), nine nasopharyngeal carcinoma biopsies (10.8%) and 13 throat washing samples (32.5%). A total of 126 isolates (76.8%) had the same amino acid sequence with the B95-8 prototype including 38 EBV-associated gastric carcinoma isolates (92.7%), 56 nasopharyngeal carcinoma biopsies (67.5%) and 32 throat washing samples (80.0%), respectively. These 126 samples, with identical amino acid sequences to the B95-8 prototype, were arranged into one group and named pattern B95-8.
Mutation of Viral Interleukin-10 Gene
A total of 18 sequence variations were found in 164 samples, including 12 non-silent mutations and six silent mutations (Fig. 3). Collectively, 19 distinct DNA sequences were identified, which encoded for 14 different BCRF1 amino acid sequences.
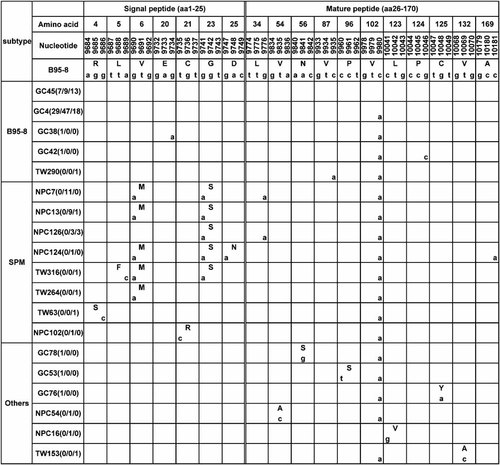
vIL-10 sequence variations in EBV-associated gastric carcinoma, nasopharyngeal carcinoma biopsies and throat washing samples from healthy donors in Northern China. Numbers across the top correspond to the amino acid positions of the gene under which the B95-8 prototype amino acid and nucleotide sequence are listed. Different patterns are noted to the left column, while the specimens showing identical sequences to each other are listed by a representative isolate in the second column. GC, EBV-associated gastric carcinoma; NPC, nasopharyngeal carcinoma; TW, throat washing samples from healthy donors. The followed numbers in the parentheses denote the amount of the identical sequences from EBV-associated gastric carcinoma, nasopharyngeal carcinoma biopsies, and throat washing samples, respectively. The small letters denote the nucleotide and the amino acids are denoted by capital letters.
Six non-silent mutations (R4S, L5F, V6M, C21R, G23S, and D25N) and one silent mutation (g9734a) were present in the signal peptide region of vIL-10. Among six non-silent mutations, three (R4S, L5F, and V6M) were found in the hydrophobic core region and three (C21R, G23S, and D25N) in polar c-region (Fig. 1). V6M and G23S were two main mutations. V6M was detected in 21 nasopharyngeal carcinoma (25.3%) and three throat washing (7.5%) samples. G23S was found in 24 nasopharyngeal carcinoma biopsies (28.9%) and five throat washing samples (12.5%). There were 32 cases (19.5%) in which a mutation in the signal peptide region was identified including 25 nasopharyngeal carcinoma biopsies (30.1%) and seven throat washing samples (17.5%), respectively. The isolates with mutations at the signal peptide region were characterized into one group and named pattern SPM.
Six non-silent mutations (V54A, N56S, P96S, L123V, C125Y, and V132A) and five silent mutations (g9776a, c9935a, c9980a, g10046c, and c10181a) were present in the mature protein of vIL-10. Among six non-silent mutations, one (V132A) was found in helix E and five in helix loops (Fig. 2). c9980a and g9776a were two main silent mutations in the mature protein. The most common silent point mutation at 9980 (c9980a) was found in 134 cases (81.7%), including 34 EBV-associated gastric carcinoma isolates (82.9%), 73 nasopharyngeal carcinoma biopsies (88.0%) and 27 throat washing samples (67.5%). Ninety-four isolates (57.3%) had only this point mutation. A silent mutation at nucleotide 9776 (g9776a) was detected in 14 nasopharyngeal carcinoma biopsies (16.9%) and three throat washing samples (7.5%).
Distribution of vIL-10 Gene Subtypes in EBV-Positive Gastric Carcinoma, Nasopharyngeal Carcinoma Biopsies and Throat Washing Samples
A total of 158 samples were classified into two types: pattern B95-8 (identical sequence to B95-8) and pattern SPM (mutation in the signal peptide). The remaining six samples, with no common mutation, were classified as “others” (Table II). The distributions of the vIL-10 types among EBV-associated gastric carcinoma group was significantly different from that in nasopharyngeal carcinoma or the healthy donor group (gastric carcinoma vs. nasopharyngeal carcinoma: P < 0.001; gastric carcinoma vs. healthy donor: P = 0.006), whereas their distribution in nasopharyngeal carcinoma and the healthy donor group was not significant different (P = 0.255).
| Subtype | EBVaGCs (n = 41) | NPCs (n = 83) | TWs (n = 40) |
|---|---|---|---|
| Pattern B95-8 | 38 (92.7%) | 56 (67.5%) | 32 (80.0%) |
| Pattern SPM | 0 (0) | 25 (30.1%) | 7 (17.5%) |
| Others | 3 (7.3%) | 2 (2.4%) | 1 (2.5%) |
- EBVaGCs, EBV-associated gastric carcinomas; NPCs, nasopharyngeal carcinomas; TWs, throat washings from healthy donors.
DISCUSSION
In all, 164 EBV-positive samples, including 41 EBV-associated gastric carcinoma isolates, 83 nasopharyngeal carcinoma biopsies, and 40 throat washing samples from healthy donors in Northern China were successfully amplified and analyzed. This study demonstrates that the vIL-10 gene is highly conserved, aside from a few point mutations, in various EBV isolates.
The protein product vIL-10 shares functional properties with hIL-10 [Vieira et al., 1991] and predominantly mediates immunosuppressive functions. The C-terminal sequences of vIL-10 gene, representing the immunoinhibitory functions (residues at 163-170, AYMTIKAR) [Gesser et al., 1997], were identical to the corresponding B95-8 sequence in all of the 164 isolates. The receptor binding sites at helices A and F and the loop between helix A and helix B (AB loop) [Zdanov et al., 1997; Yoon et al., 2005] were conserved in all EBV-associated gastric carcinoma, nasopharyngeal carcinoma and throat washing samples, except for GC78 (N56S) and NPC54 (V54A, Fig. 2). It has been demonstrated that a single amino acid, isoleucine, at position 87 of hIL-10 is required for its immunostimulatory function [Ding et al., 2000]. Substitution of alanine with isoleucine in vIL-10 at the corresponding position 98 alters its receptor binding affinity and spectrum of biological activities and contributes to ineffective host immune response [Yoon et al., 2005]. In all EBV-associated gastric carcinoma, nasopharyngeal carcinoma and throat washing cases, this position was highly conserved. The overall homology of the EBV-associated gastric carcinoma, nasopharyngeal carcinoma and throat washing sequences exceeded 98.8%. A total of 126 isolates (76.8%) had the same amino acid sequence as the B95-8 prototype. It is thought that the high conservation of vIL-10 gene illustrates the relevance for the virus to evade host immunity, because vIL-10 mimics the activity of hIL-10.
According to the signal hypothesis, the signal peptide is eventually cleaved by signal peptidase to form a mature protein. There were 32 cases (19.5%) in which amino acid changes in the signal peptide region was identified. While, only six isolates (4.0%) had amino acid changes at the mature protein. Compared with the mature protein, the signal peptide was prone to harbor more variation.
Signal peptides typically consist three domains, a positively charged (n-) region, a hydrophobic core (h-) region, and a polar carboxy-terminal (c-) region [Perlman and Halvorson, 1983; von Heijne, 1985] (Fig. 1). The hydrophobic core region is the most essential part required for targeting and membrane insertion [von Heijne, 1985]. Among the three mutations which located at the hydrophobic core (h-) region, the consensus mutation V6M and one sporadic mutation L5F were changed among the hydrophobic amino acid residues, while another sporadic mutation R4S mutated from charged Arg to polar Ser. R4S mutation had been also observed in the EBV positive cell line, EB1 [Kanai et al., 2007]. The polar c-region often contains helixbreaking proline and glycine residues as well as small uncharged residues in positions -3 and -1 that determine the site of signal peptide cleavage [von Heijne, 1990]. Acceptable cleavage domains conform to the following rules: the residue at position -1 from the cleavage site must be small; the residue at position -3 must not be aromatic, charged, or large polar residues; and there must be no Pro residue in the region between -3 and -1 position [Perlman and Halvorson, 1983; von Heijne, 1983]. Among the three mutations which located at the polar c-region, the consensus mutation G23S at position -3 and the sporadic mutation D25N at position -1 followed these rules, while another sporadic mutation C21R changed from positively charged Arg to polar Cys at -5. Previous studies showed that substitution of a charged amino acid (Arg for Cys) in the signal peptide hydrophobic core of human preproparathyroid hormone (PPTH) causes dominant secretory dysfunction [Karaplis et al., 1995], and a positively charged Arg for polar Gly substitution at -4 in polar c-region of IL-10 affects the efficiency of protein translocation and signal peptide cleavage resulting in lower levels of IL-10 protein secretion [Whittington et al., 2003]. Considering the similarities among the structures of different signal peptides, it can be deduced that the mutations of the amino acid in the peptide region of vIL-10 may also influence its functions, such as peptide cleavage and protein secretion. Therefore, the function of the SPM variant is valuable to be determined in further study.
Among the EBV isolates from Northern China in this study, the amino acid sequences of major isolates were conserved and identical to the B95-8 sequence. These samples were classified into one group pattern, B95-8. This pattern was the dominant type in the study and was present in 126 samples (80.8%), including 38 EBV-associated gastric carcinoma isolates (92.7%), 56 nasopharyngeal carcinoma biopsies (67.5%), and 32 throat washing samples (80.0%). The major EBV-associated gastric carcinoma and throat washing isolates (21/22) in a Japanese study had the same amino acid sequences as B95-8 [Kanai et al., 2007].
Compared to the EBV-associated gastric carcinoma isolates, the EBV-positive nasopharyngeal carcinoma biopsies and throat washing samples exhibited more mutations. Pattern SPM (mutation in the signal peptide) existed only in the EBV-positive nasopharyngeal carcinoma and throat washing isolates, including 25 nasopharyngeal carcinoma biopsies (30.1%) and seven throat washing samples (17.5%), not in EBV-associated gastric carcinoma isolates. Thus pattern SPM seems more relevant in the EBV-positive nasopharyngeal and laryngopharyngeal epithelial cells. Although distributions of the vIL-10 type in nasopharyngeal carcinoma and throat washing samples was not significantly different (P = 0.255), pattern SPM was more common in nasopharyngeal carcinoma biopsies (25/83, 30.1%) than in throat washing samples (7/40, 17.5%). Furthermore, most (21 out of 25) nasopharyngeal carcinoma biopsies with the SPM pattern carried two consensus signal peptide mutations (V6M and G23S), while only two of the seven throat washing samples did. These results suggest that the SPM pattern is preferentially found in nasopharyngeal carcinoma biopsies. The nasopharyngeal carcinoma isolates harbored more variation than the other two groups, which is in accordance with results from the analysis of a polymorphism of EBER in Northern China [Wang et al., 2010]. It is deduced that a substrain of EBV with the SPM pattern of vIL-10 gene preferentially infects nasopharyngeal carcinoma and a susceptibility to a particular EBV isolate in the nasopharynx may exist during the formation of nasopharyngeal carcinoma. However, it is difficult to examine matched peripheral blood lymphocytes (PBLs) or throat washing samples with tumor biopsies due to the current unavailability of these samples. There is no report on the polymorphism of the vIL-10 gene in southern China, the area in which nasopharyngeal carcinoma is endemic. Thus, whether this new SPM pattern is specific geographically or associated preferentially with nasopharyngeal carcinoma remains to be studied.
In conclusion, this study indicates that v-IL10 is a conserved gene, especially in its functional domains. Pattern B95-8 is the commonest in this region. Pattern SPM seems to be associated to EBV-positive nasopharyngeal and laryngopharyngeal epithelial cells. The observation that the vIL-10 gene is highly conserved demonstrates its important role in maintaining the basic biological activity of the virus.
Acknowledgements
Authors thank Rebecca Leeman-Neill, PhD, MD (University of Pittsburgh, Pittsburgh, PA, USA) for her kind review of this manuscript.




