Molecular characterization of hepatitis C virus genotype 4 sequences in HIV-coinfected patients from Argentina
Abstract
The prevalence of hepatitis C virus genotype 4 (HCV-4) is increasing in different parts of the World but in Latin America the data are still scarce. We aimed to characterize HCV-4 isolates from 383 HIV-coinfected patients in Argentina. Sequence analyses were based on the non-structural 5B region of HCV. Results from 18 patients indicated a genetic heterogeneity that involved three genotype 4 subtypes. Sequences were ascribed to subtype 4d (67%), 4a (22%), and 4m (11%). In spite of different sources of transmission were defined among patients, no statistical association was found with the genotype 4 subtype. The scenario is also compatible with multiple importation of the epidemic and there is no evidence for transmission-specific clusters or network-like transmission of HCV-4. This HCV-4 does not represent a recent introduction in Argentina, it circulates in all transmission groups and its presence is increasing among HIV-infected patients. J. Med. Virol. 83:935–940, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis C virus (HCV) and human immunodeficiency virus (HIV) share the same routes of transmission, which explains the high rate of HCV and HIV coinfection.
For HCV, at least seven major genotypes and more than 67 subtypes have been identified [Simmonds et al., 2005; Kuiken and Simmonds, 2009]. Research to relate specific genotype groups to the outcome of infection has been done. Some associate genotypes with a predominant transmission route [Ramalho et al., 2000] or a particular interferon resistance profile [Maekawa and Enomoto, 2009]. Likewise, it has been observed that the genotypes have been related with a particular geographic distribution. Some seem to have spread worldwide (genotypes 1a, 1b, 2a, 2c, 3a), while others have been found in more restricted regions only (genotypes 4, 5a, and 6a) [Bostan and Mahmood, 2010].
HCV genotype 4 (HCV-4) is common in the Middle East and in Africa, where it is responsible for more than 80% of HCV infections, and has recently spread to several European countries [de Bruijne et al., 2009; van de Laar et al., 2009; Eriksen et al., 2010; Vogel et al., 2010]. There is considerable diversity among isolates and several subtypes have already been identified, even in the same geographic region where this subtype prevails [Li et al., 2009]. Although HCV-4 is responsible of approximately 20% of the 170 million cases of chronic hepatitis C in the world, it has simply not been the subject of widespread research; therefore, the features of this genotype and management strategies for patients infected with this genotype are still insufficient when comparing with genotypes 1, 2, and 3 [Kamal and Nasser, 2008].
In Argentina, two previous studies have shown that HCV-4 was only found in a minor proportion of HCV mono-infected patients [Picchio et al., 1997; Alfonso et al., 2001] where the most prevalent genotype is type 1, with the presence of other genotypes [Quarleri et al., 2000]. The scarce sequence data related to HCV-4 prompted us to characterize at genomic level newly genotype 4 variants isolated from Argentinean HIV-1 coinfected patients and to investigate their evolutionary relationships with other genotype 4 subtypes intending to increase the understanding of the spread of HCV-4.
MATERIALS AND METHODS
Patients
The present study included 383 HCV-HIV coinfected patients who have been diagnosed positive for HCV antibodies by an enzyme immunoassay and serum HCV RNA. They are HIV-infected patients attending at different hospitals in Buenos Aires city. Blood samples were obtained from these patients for regular monitoring from May 2008 to May 2010. This study was approved by the Huesped Foundation Ethics Committee and consents were obtained for all participating patients.
Hepatitis C Virus Genotyping
HCV viral load (Bayer VERSANT® HCV RNA 3.0 Assay) and HCV genotyping (Inno-LiPA HCV II, Innogenetics, Ghent, Belgium) of 383 HIV-HCV coinfected patients were determined. These procedures were carried out immediately prior the patients started their anti-HCV pegylated-interferon plus ribavirina therapy. All patients were under antiretroviral therapy and their mean ± SD HIV viral load were 135 ± 43 copies/ml (VERSANT® HIV-1 RNA 3.0 Assay).
The resulting molecular epidemiology distribution pattern (genotype 1, 73.4% of isolates; genotype 2, 2.1%; genotype 3, 17.5%; genotype 4, 3.7%; mixed genotypes, 3.3%) squared with the prevalence previously reported in Argentinean HIV-infected patients [Quarleri et al., 2007; Bolcic et al., 2008]. Four additional genotype 4 isolates (Patient 15–Patient 18) previously found in a study carried out by the authors [Bolcic et al., 2008] but not characterized at NS5B genomic level were added to the present phylogenetic study as well as to investigate any possible epidemiological linkage among HCV genotype 4 strains. Thus, 18 sequences from the NS5B region of the viral genome were obtained.
HCV RNA Isolation, Reverse Transcription, NS5B PCR, and Sequencing
The viral RNA extraction, reverse transcription, and amplification were performed as follows: RNA was extracted from 100 µl of plasma using a solution of phenol and guanidine isothiocyanate (TrizolTM LS Reagent; Gibco BRL, Gaithersburg, MD), according to the manufacturer's instructions. The final product was dissolved in 20 µl of RNase-free water. Reverse transcription was performed with MMLV reverse transcriptase and random hexamers. The NS5B PCR protocol was based on that previously reported by [Laperche et al., 2005]. Briefly, cDNA was amplified by a heminested PCR based on primers PR3 and PR4 in the first round, followed by PCR with primers PR3 and PR5 in the second round obtaining a 401-bp product. The PCR product encompasses nucleotide positions 8245 and 8645 based on the numbering system of Choo et al. [1991]. The amplified products were electrophoresed through a 1.5% agarose gel and visualized by ethidium bromide staining. The DNA of each strain obtained from the purified PCR products (Quick Spin; Qiagen, Benelux, Belgium) was directly subjected to double strand sequencing with dye-labeled dideoxy terminators by PCR with primers PR3 and PR5 (ABI PRISM 3100 automated sequencer, Applied Biosystems, Foster City, CA).
HCV Phylogenetic Analysis
DNA sequences were edited and assembled using Sequencer software v.4.10.1 (Gene Codes). Reference sequences for the various HCV geno/subtypes were obtained from GenBank and European Hepatitis C database [Kuiken et al., 2005; Combet et al., 2007] including sequences for the different subtypes of genotype 4. Three previously reported Argentinean HCV genotype 4 sequences were also included [Alfonso et al., 2001] (Fig. 1).
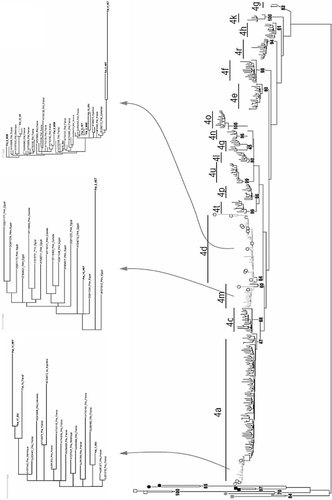
Maximum likelihood phylogenetic tree of the NS5B region showing the genotype 4 distribution of the Argentinean isolates (open circles). Three genotype 4 Argentinean sequences (AF308573; AF308574; AF308575) from a previous study [Alfonso et al., 2001] are also shown. Outgroup sequences are indicated with closed black (genotype 1), gray (genotype 3) or open (genotype 2) squares, black circles (genotype 6) and gray closed circles (genotype 5). The clades into which Argentinean sequences were clustered are shown in detail; route of transmission is defined for each one. GenBank accession number and country sampling from HCV genotype 4 references sequences are indicated. Branch lengths are proportional to the number of nucleotide substitutions per aligned site (bars = 0.01 substitutions).
Multiple sequence alignment was achieved by the Mafft program [Katoh et al., 2002, 2009].
Two datasets were input into phylogenetic analyses. The first one, including 407 sequences was analyzed by Likelihood and Parsimony techniques. The second one, which included 117 sequences, was analyzed by Bayesian methods, as larger datasets precluded convergence of the Markov processes. For the probabilistic analyses, the model of sequence evolution was inferred by MrAIC [Nylander et al., 2004]. The model with the best fit the data was GTR+I+G. Maximum likelihood trees were inferred with PhyML version 3.0 [Guindon and Gascuel, 2003] using a BIONJ starting tree that was reordered by SPR for searching the tree space. Bayesian analyses were performed with the program MrBayes program [Huelsenbeck and Ronquist, 2001]. The analyses were run independently two times using Metropolis-coupled Markov Chain Monte Carlo (MCMC) enhance the tree-climbing capabilities of the Markov chains [Huelsenbeck and Ronquist, 2001]. The clades' posterior probabilities were estimated as the proportion of samples presenting any particular clade. For the Parsimony analyses, the TNT program was carried out [Giribet, 2005; Goloboff et al., 2008]. Tree searches were performed as described elsewhere [Dilernia et al., 2008]. Shortly, cycles of random addition sequences plus tree-bisection-reconnection followed by ratchet, tree-drift and tree fussing until no further improvement of tree lengths was observed.
All HCV genomic sequences, sequence alignments, and phylogenetic trees are available upon request from the authors. The genotype 4 reference strains included in the phylogenetic tree are named with GenBank accession numbers, genotype/subtype and sampling country when possible (Fig. 1).
Statistical Analysis
Differences between data groups were analyzed with the independent t-test and Chi-square test (“SISA-Binomial” software packages [Uitenbroek, 1997]). The P-values (two-tailed) <0.05 were considered statistically significant.
RESULTS
The distribution of HCV genotypes among 383 HIV-infected patients showed that 14 samples, turned out to be genotype 4 exhibiting an statistically significant (P = 0.01) fourfold increase of its prevalence among HIV-infected patients in a short period of time (2005–2009) considering previous HCV molecular epidemiology study among HIV-coinfected patients [Quarleri et al., 2007].
Molecular Characterization of Argentinean HCV Genotype 4 Isolates
The distribution of HCV-4 according to epidemiological characteristics of the patients showed statistical differences only against genotype 2 regarding age (45.5 ± 1.7 vs. 65.0 ± 3.2, respectively; P < 0.0005) and to viral load against genotype 1 (5.49 ± 0.27 vs. 5.85 ± 0.33, respectively; P < 0.003). The HCV-4 was found more frequently in men (56%) than in women (Tables Ia and Ib).
| HCV genotype | 1 | 2 | 3 | 4a | Mixed |
|---|---|---|---|---|---|
| N = 383 (%) | 281 (73.4) | 8 (2.1) | 67 (17.5) | 14 (3.6) | 13 (3.4) |
| Mean HCV-VL (log IU/ml) | 5.85 | 5.21 | 5.3 | 5.49 | 5.39 |
| Female/male | 91/190 | 1/7 | 23/44 | 7/7 | 1/12 |
| Mean age (years) | 42.7 | 65 | 43.4 | 45.5 | 41 |
- N, number of patient.
- a The four additional patients infected by HCV genotype 4 (Patients 15–18) depict the following basal characteristics: mean HCV-VL (log IU/ml): 5.82; female/male ratio: 1:3; mean age (years): 39.6.
| Characteristics | Patients, N (%) |
|---|---|
| All patients | 18 (100) |
| Gender | |
| Male | 10 (56) |
| Female | 8 (44) |
| Age (years) | |
| Median (IQR) | 43.5 (39–45) |
| Route of transmission | |
| Injecting drug use | 6 (33) |
| Blood transfusion | 1 (6) |
| Sexual contact | |
| Heterosexual | 6 (33) |
| MSM | 4 (22) |
| Unknown | 1 (6) |
| Subtypes | |
| 4a | 4 (22) |
| 4d | 12 (67) |
| 4m | 2 (11) |
- IQR, interquartile range; MSM, men having sex with men.
The resulting data from the 18 HCV-4 strains that were subjected to direct sequencing and subsequent phylogenetic analysis performed by maximum likelihood (Fig. 1) as well as by maximum parsimony and Bayesian analysis (data not shown) demonstrated the clustering of all these sequences with the appropriate reference sequences after using BLAST and each closest match for genotype 4 subtypes was retrieved. Sequences were ascribed to subtype 4d (67%), 4a (22%), and 4m (11%). The GenBank accession numbers for the sequences reported in this work are HQ438947–HQ438964.
All patients were ethnic Argentinean. Different sources of transmission were defined among patients. Those four infected by genotype 4a were two injecting drug users (IDU), a female sex worker and a patient who received a blood transfusion. Regarding the 12 patients infected by genotype 4d, 4 were IDU and other 4 men who have sex with men (MSM). The remaining referred heterosexual behavior but one declared unknown risk factors. Finally, those isolates ascribed to subtype 4m were identified in two patients with sexually transmitted risk of infection (Tables Ia and Ib). The patients, who attended at different hospitals, denied having traveled to endemic areas. No statistical association was found among source of infection and the subtype of genotype 4 (P > 0.05, Chi-square test).
Comparison With HCV Genotype 4 Published Sequences
In order to compare the worldwide transmission routes with the Argentinean isolates, further phylogenetic analysis of the HCV-4 Argentinean strains along with other from different parts of world by using available databases [Kuiken et al., 2005; Combet et al., 2007] was carried out. This analysis did not reveal a segregation of the Argentinean sequences with those characterized in a particular country. In line, the 4d (Pat 3, 4, 5, 6, 7, 8, 9, 10, 12, 13, 16, 18) and 4a (Pat 1, 11, 14, 17) strains characterized from HIV-coinfected appeared in the NS5B phylogenetic tree associated with HCV-4d (AF308574 and AF308575) and HCV-4a (AF308573) strains previously identified from Argentinean patients without HIV co-presence [Alfonso et al., 2001] as well as with other strains from France and Egypt. The remaining two HCV-4 isolates (Patients 2 and 15) were classified as subtype 4m closely related with Egyptian isolates (Fig. 1).
DISCUSSION
The HCV-4 scenario among HIV-coinfected patients in Argentina is compatible with multiple importation of the epidemic and there is no evidence for transmission-specific clusters or network-like transmission of HCV-4.
Overall the data of genotypic specific prevalence of HCV-4 coupled with the phylogenetic comparison suggests that the genotype 4 does not represent a recent introduction in Argentina, it circulates in all transmission groups and its presence is increasing among HIV-infected patients. These facts reject the hypothesis that all of the HCV-4 isolates had a common source.
It should be observed that the HCV-4 did not display a common route of infection as was reported in Spain [Sanchez-Quijano et al., 1997] or associated with a particular epidemiological profile [de Bruijne et al., 2009; van de Laar et al., 2009; Eriksen et al., 2010; Vogel et al., 2010]. However, considering that the number of patients infected with HCV genotype 4 is reduced, the identification of network may have been hampered.
The genotype 4 subtypes infecting the patients coinfected with HCV and HIV varied, and the 4d subtype dominated (Tables Ia and Ib). The HCV subtype 4d, 4a, and 4m sequences from coinfected patients did not cluster together, and there was no indication of a connection between these patients. But the scarce number of patients included within each of them precludes any further conclusions. The coexistence of diverse subtypes of this genotype is described for regions which display a high prevalence of genotype 4 infections such as Middle East and in northern and central Africa [Smith et al., 1997]. The prevalence in Europe has increased due to immigration of HCV carriers [de Bruijne et al., 2009; van de Laar et al., 2009; Eriksen et al., 2010; Vogel et al., 2010]. In Latin America the information is still scarce. The observed polyphyletic cluster of HCV-4 in Argentina appears to represent its spillover into HIV-coinfected patients reporting IDU and MSM as more frequent associated risk transmission. Since infections with HCV-4 are difficult to treat successful therapy against HCV-4 will increasingly be needed.
These data may be useful for the long-term planning of control of HCV infection in Argentina where a low prevalence of this HCV genotype was reported up to now. Future detection of genotype 4 in this area may deserve special attention.




