Impact of universal vaccination on intrafamilial transmission of hepatitis B virus†
Shu-Chi Mu and Gen-Ming Wang contributed equally to this work.
Abstract
To control hepatitis B virus (HBV) infection, a nationwide vaccination program was launched in 1984 and resulted in a significant reduction in the rate of persistent infection of children. However, the relative contribution of vaccination to the intrafamilial clustering of HBV infection remains unclear. The rate of intrafamilial HBV transmission in vaccinated children was investigated. Eighty-four sera from vaccinated children were enrolled and HBV serum markers were determined. The modes of intrafamilial HBV transmission were investigated by history taking and serological assay, and confirmed by genotyping and phylogenetic analysis. The results showed 66 (78.6%) vaccinated children born to hepatitis B surface antigen (HBsAg)-negative parents were HBsAg-negative. Eighteen vaccinees were born to HBsAg-positive parents; four (21.4%) of the children were HBsAg-positive. According to the parents' HBsAg status, three patterns of HBsAg-positive parents were identified. Serological analysis showed that three of 15 children born to HBsAg-positive mother (pattern I) and one of two children born to HBsAg-positive father became infected (pattern II). The remaining one child was HBsAg negative with both parents positive for HBsAg (pattern III). Genotyping and phyogenetic analysis confirmed the mode of intrafamilial transmissions. Sequence analysis of S and pre-S genes showed that HBV isolates of HBsAg-positive vaccinees were variants; no G145R but G145A and other substitutions were found. In conclusion, this small study showed that both maternal and paternal transmissions are important of the intrafamilial spread of HBV infection. In addition, the introduction of HBV vaccination has resulted in a reduction of intrafamilial transmission, but a study of a large population is needed. J. Med. Virol. 83:783–790, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection is a major health problem worldwide, affecting approximately 350 million individuals. The clinical outcomes of chronic HBV infection include the inactive carrier state, chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [Seeger and Mason, 2000; Kao and Chen, 2002]. Clustering of chronic HBV infection within the family is common in endemic areas [Szmuness et al., 1973; Sung and Chen, 1978], and perinatal transmission has been shown to be the major route of transmission [Stevens et al., 1975; Sung and Chen, 1980]. Horizontal transmission of the virus in early childhood as a result of close family contact is also important [Szmuness et al., 1973; Lok et al., 1987; Dumpis et al., 2001].
Taiwan is a hyper-endemic country for HBV infection and, previously, as many as 15–20% of general population were chronic HBV carriers [Chen et al., 1996]. The majority of chronic HBV carriers became infected early in life while living in endemic areas, especially before the age of 2 years [Stevens et al., 1975; Beasley et al., 1982; Hsu et al., 1986]. Both perinatal transmission and childhood intrafamilial horizontal infection are common in Taiwan [Lin et al., 2005; Chen, 2009]. To control HBV infection, a nationwide vaccination program was launched in 1984 [Chen et al., 1987]. All infants received three to four doses plasma or recombinant HBV vaccines. In addition, infants born to HBeAg-positive mothers received 0.5 ml of hepatitis B immunoglobulin within 24 hr after birth. This program resulted in a significant reduction in the rate of persistent infection of children and the occurrence of childhood hepatocellular carcinoma and fulminant hepatitis in Taiwan [Chang, 2006; Chen, 2009].
However, the influence of universal vaccination on intrafamilial HBV infection had not been studied. The aims of this study were to evaluate the changes in intrafamilial transmission after the introduction of HBV vaccination. In addition, possible routes of transmission and risk factors for intrafamilial transmission, and the impact of vaccine escape mutants on intrafamilial transmission were investigated.
PATIENTS AND METHODS
Vaccination Schedule
All children investigated received 3 doses of recombinant vaccine H-B-Vax II (5 µg/0.5 ml; Merck Sharp & Dohme, Rahway, NJ) or Engerix-B (20 µg/1 ml; SmithKline Beecham, Rixensart, Belgium) administered at <1 week, 1 month, and 6 months of age. In addition, their mothers were screened for HBsAg and hepatitis B e antigen (HBeAg) if HBsAg was found. Hepatitis B immunoglobulin (HBIG) 0.5 ml (100 IU) was given within 24 hr after birth to newborns of HBeAg-positive or high titer of HBsAg (reciprocal titer >1:2560 by the reverse passive hemagglutination test) carrier mothers. The immunization history was checked by examining the vaccination card and history taking from the parents.
Subjects and Their Families
Eighty-four vaccinated children from 82 families were recruited at random during June, 2007 to September, 2009 in the Shin Kong Wu Ho-Su Memorial Hospital. Most of these children had some medical conditions such as IgE-related allergy, respiratory tract infection or gastroenteritis which required medical consultation. The history of familial clustering of HBV infection and hepatitis B vaccination were recorded. Consent for participating in the study was filled by the parents. In addition, sera of the parents of two families with evidence of clustering HBV carriers were collected. The serum samples obtained from all patients were tested for hepatitis B surface antigen (HBsAg), hepatitis B surface antibody (anti-HBs), and hepatitis B core antibody (anti-HBc). Serum samples were divided into aliquots and kept at −80°C until testing.
Serological Markers
HBsAg, anti-HBs, and anti-HBc were tested with commercial kits (Elecsys HBsAg, anti-HBs, and anti-HBc, respectively; Roche Diagnostics, Mannheim, Germany).
Extraction of Serum HBV DNA
Serum viral DNA was extracted from 200 µl serum by using commercial kits (QIAamp DNA Blood Mini Kit, Qiagen Inc., Valencia, CA). The extracted DNA was used for amplification and direct sequencing of S gene as described previously (Chen et al., 2004; Mu et al., 2009).
Quantification and Genotyping of HBV Viral DNA
HBV DNA was quantified and genotyped by real-time PCR based on the LightCycler hybridization probes assay system. The primers covered nucleotide positions 1261–1279 and 1600–1580; the anchor probe was nucleotide position 1552–1576, and the sensor probe was nucleotide position 1533–1550. The plasmid pHBV-48 (derived from subtype adw2) was used for generating the copy number standard curves for viral load quantitation. The detail nucleotide sequences and procedures were described previously [Yeh et al., 2004]. Real-time PCR monitoring was achieved by measuring the fluorescence at the end of the annealing phase for each cycle. The measurement was performed by using the LightCycler analysis software 3.5 (Roche Diagnostics Applied Science). The sensitivity of this method was 102 copies/ml.
Amplification and Sequencing of the Surface, and Pre-S Genes
The segments of surface antigen (549 bp, nucleotide positions 247–795) and pre-S (564 bp, nucleotide positions 2828–176) DNA were amplified by nested PCR and sequenced as described previously [Chen et al., 2006; Mu et al., 2009]. The forward primers of S gene were changed into S-3, 5′- TCCTAGGACCCCTGCTCGTGTTAC -3′ for outer set and S-5, 5′- TCTAGACTCGTGGTGGACTT -3′ for inner set. To avoid false positivity of PCR, precautions described by Kwok and Higuchi [1989] were followed strictly.
Alignment and Phylogenetic Analysis
Alignment analysis was performed using the Biology WorkBench 3.2—CLUSTALW software program (http://workbench.sdsc.edu) [Thompson et al., 1994].
Phylogenetic analysis was used to identify the mode of transmission in families infected with the same HBV genotype. Nucleotide sequences of the four genotypes (A-D) were obtained from the GenBank, European Molecular Biology Laboratory, and DNA Data Bank of Japan databases. The accession numbers of 22 sequences are indicated in Figure 2. A phylogenetic tree was constructed by using neighbour-joining method component of the Molecular Evolutionary Genetics Analysis (MEGA) 3 program [Kumar et al., 2004] on the basis of the nucleotide sequences of the amplified S gene of the HBV genome. Genetic distances were estimated by using the 6-parameter method, and phylogenetic trees were constructed by the neighbor-joining method [Saitou and Nei, 1987]. To confirm the reliability of the phylogenetic tree analysis, bootstrap re-sampling and reconstruction were performed 500 times.
Ethical Considerations
The study was approved by the Ethical Committee of the Shin Kong Wu Ho-Su Memorial Hospital, and the sera samples were collected after receiving consent from the patients or their parents if the patients were minors. Patients with HBV infection and their parents were informed about positive results.
Statistical Analysis
Data were analyzed by chi-square test and Fisher's exact test when appropriate. All of the tests of significance were two-tailed with a P value less than 0.05.
RESULTS
The Prevalence of HBsAg Positivity
Since this is a preliminary study, in order to obtain the record of the histories of familial clustering of HBV infection and hepatitis B vaccination, samples of the children born in one hospital were obtained. A total of 84 vaccinated children (40 females and 44 males) from 82 families with parental serological markers were selected. All families investigated are single-child families except two; siblings of the children from these two families were also recruited. The vaccination coverage rate in the population of subjects in this study was 100%. The age of the vaccinees ranged between 1.1 to 13.3 years (average 5.6 years). The overall prevalence of HBsAg in vaccinated children was 4.8%. In addition, these subjects were classified into two groups according to the HBsAg status of their parents (Table I). Group I included 66 (78.6%) children born to HBsAg-negative parents. A total of 18 (21.4%) vaccinated children (nine males and nine females) from 17 families with evidence of HBsAg-positive parents were classified into group II. According to the parents' HBsAg status, three patterns could be identified (Table I). Of the 17 families, 14 were with an HBsAg-positive mother (pattern I) and two were co-positive HBeAg at delivery, two were with an HBsAg-positive father (pattern II), and the rest was with HBsAg-positive parents and mother was HBeAg-copositive at delivery (pattern III). All children in group I were HBsAg-negative. Conversely, in group II, the rates of HBsAg-positive in parents and children were 52.9% (18/34) and 22.2% (4/18), respectively. The rates of HBsAg-positive in children of all three representative patterns were 3/15 (20%), 1/2 (50%), and 0/1 (0%), respectively.
| HBsAg status | No. of couples | No. of HBsAg-positive children/no. of children | ||
|---|---|---|---|---|
| Male | Female | Total | ||
| Group I: HBsAg negative parents | 65 | 0/35 | 0/31 | 0/66 |
| Group II: HBsAg positive parents | ||||
| Pattern I, HBsAg-positive mothera, HBsAg-negative father | 14 | 2/9 | 1/6 | 3/15 |
| Pattern II, HBsAg- negative mother, HBsAg- positive father | 2 | 0/0 | 1/2 | 1/2 |
| Pattern III, motherb and father both HBsAg- positive | 1 | 0/0 | 0/1 | 0/1 |
| Total | 17 | 2/9 | 2/9 | 4/18 |
- a Two were positive with HBsAg and HBeAg at delivery.
- b Mother was positive with HBsAg and HBeAg at delivery.
The Relationship of HBV Infection to Age, Sex, Anti-HBs Positivity, and an HBsAg Positive Parent
The association of HBsAg positivity with selected factors was investigated and shown in Table II. There was no significant difference in terms of children aged less than six years old (P = 0.324), gender (P = 1.00), or anti-HBs positivity (P = 0.343). In contrast, the rate of HBsAg-positive offspring with HBsAg-positive parents was significantly higher than those with HBsAg-negative parents (22.2% vs. 0.0%, P < 0.005).
| Factor | Total no. (%) | No. (%) of HBV infection | P value |
|---|---|---|---|
| Age | |||
| <6 years | 46 (54.8) | 1 (2.2%) | 0.324 |
| >6 years | 38 (45.2) | 3 (7.9%) | |
| Sex | |||
| Female | 40 (47.6) | 2 (5%) | 1.00 |
| Male | 44 (52.4) | 2 (4.5%) | |
| Anti-HBs | |||
| Negative | 40 (47.6) | 3 (7.5%) | 0.343 |
| Positive | 44 (52.4) | 1 (2.3%) | |
| HBsAg positive parents | |||
| Presence | 18 (21.4) | 4 (22.2%) | <0.005 |
| Absence | 66 (78.6) | 0 (0%) | |
Genotyping and Phylogenetic Analysis
The results of history taking and serological assay showed that four of 84 children had evidence of intrafamilial transmission of HBV, three children were from maternal transmission and their mother was HBeAg negative at delivery of the newborn; one was from paternal transmission (Fig. 1). In order to trace the possible modes of transmission of intrafamilial clustering of HBsAg carriers, genotyping and phylogenetic analysis of the S gene of HBV genome were used to identify. Two of four families had sufficient serum for isolating viral DNA. The results of genotyping showed that the daughter and the father of family 1 were genotype B (pattern II); the son and the mother of family 2 were genotype C (pattern I); the titer of HBV isolates were 1.49 × 106, 1.44 × 106, 3.23 × 106, and 2.28 × 107 copies/ml, respectively (Fig. 1). In order to confirm intrafamilial transmission, the phylogenetic analysis of the S gene was done, and the results suggested that the father of family 1 might have served as the source of infection for his child, and the mother of family 2 as the infectious source of her child (Fig. 2). The HBV titer and genotype of the other two HBsAg-positive children (Fam3-Daughter and Fam4-Son) were also determined and shown in the Figure 1.
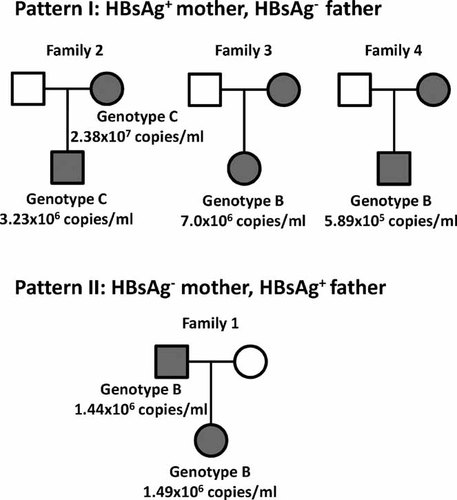
Family trees for four families with clustering infection of hepatitis B virus (HBV). The modes of transmission were confirmed by identifying the concordant HBV genotypes between HBsAg positive children and their parents in two families. Squares denote male sex; circles denote female sex; gray color indicates HBsAg positive; and white color indicates HBsAg negative.
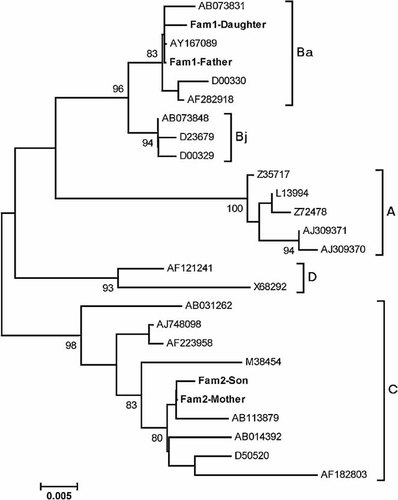
Phylogenetic tree analysis of the S sequences of HBV in two families (family 1 and 2) compared with HBV strains representative of genotype A-D. HBV strains were retrieved from DDBJ/EMBL/GenBank and their accession numbers are indicated. Pattern I: family 2 (Fam2-Mother and Fam2-Son) with an HBsAg-positive mother. Pattern II: family 1 (Fam1-Father and Fam1-Daugther) with an HBsAg-positive father. Genetic distances were estimated by the six-parameter method and phylogenetic trees were constructed by the NJ method. Bootstrap values are shown along main branch (values of 80% and higher are shown). The lengths of the horizontal bars indicate the number of nucleotide substitutions per site.
Mutations at the Amino Acid Level in the S Gene Region
To elucidate whether mutations in the “a” determinant (aa 122–148) and MHR (major hydrophilic region, aa 99–169) of HBsAg caused vaccine failure and intrafamilial transmission, the sequences of S gene were analyzed. Amino acid sequences of S gene were shown in Figure 3 where members of family 1, 3, and 4 were serotype adw, and members of family 2 were serotype adr. Two substitutions outside “a” determinant (I198M and F200Y) were detected in the isolates of family 1; the sequences of S gene are the same between daughter and father. Six substitutions (I110L, S113T, K160R, Y161F, I198M and F200Y) inside MHR and two were within the “a” determinant (T126I and T143S) were detected in the isolates of family 2. An additional mutation G145A was found in the son of family 2. The S sequences of the other two HBsAg-positive children (Fam3-Daughter and Fam4-Son) were also determined. One change (I110L in Fam3-Daughter) within the MHR and one or two substitutions outside the MHR (I198M in Fam3-Daughter; I198M and F200Y in Fam4-Son) were detected.

Amino acid sequences of HBV partial surface gene (amino acid 81–210) encompassing the “a” determinant (aa 122–148) in the four HBsAg positive children and their parents. Dot indicates identity to the reference sequence of genotype Ba/serotype adw2 isolate (D00330).
Sequence Alignment of the Pre-S Gene
To elucidate whether variation in the pre-S region caused vaccine failure, the sequences of pre-S gene were analyzed. The nucleotide sequence of pre-S amplicons from four infected children were more diverse (Fig. 4). Notably, amino acid conservation within B-cell (aa 12–32, aa 19–26, aa 72–78 and aa 94–105) and T-cell (aa 12–21 and aa 21–30) epitope of the pre-S1 region and the pre-S2 region (aa 1–6 for B cell and aa 21–30 for T cell) was found among these isolates. On the other hand, there were many differences within T-cell (aa 29–48 and aa 94–117) and B-cell (aa 27–35, aa 37–45, aa 41–53, and aa106–117) epitopes of the pre-S1 region, and B- (aa 3–15, aa 13–24, and aa 38–48) and T-cell epitope (aa 29–48) of the pre-S2 region (Fig. 5).

The nucleotide sequences of the pre-S gene in the four HBsAg positive children. Dot indicates identity to the reference sequence of genotype Ba/serotype adw2 isolate (D00330). Positions of nucleotide sequences of the pre-S region, the start codon of pre-S1 and pre-S2 genes are indicated.

The amino acid sequences of the pre-S gene in the four HBsAg positive children. Dot indicates identity to the reference sequence of genotype Ba/serotype adw2 isolate (D00330). The conservation of each amino acid is indicated under the sequence as follow: asterisks indicate as full conservation; colons, strong conservation; triangles, weak conservation; and spaces, no conservation. B-cell epitopes are showed in box; T-cell epitopes are showed underline; conversed epitopes are shaded.
DISCUSSION
The results of this study indicate that the rate of HBsAg-positive in vaccinated children collected in one hospital was 4.8%, which was lower than that in the general population of Taiwan before vaccination [Ni et al., 2007; Chen 2009]. In addition, it was found that universal vaccination blocked significantly the transmission of HBV infection in vaccinated children born to HBsAg-negative parents. It also reduced the rate of HBsAg-positivity to 22.2% in vaccinees born to HBsAg-positive parents, which was significantly lower than that of a previous observation in a similar population before vaccination (22.2% vs. 77.8%, P < 0.001) [Lin et al., 2005]. Statistic analysis revealed that the presence of HBsAg- positive parents was associated significantly with the infection of HBV in vaccinated children. This fact strongly suggested that HBV infection in parents indeed increased the frequency of intrafamilial HBV infection, irrespective of a maternal or paternal mode of transmission. It is generally considered that perinatal transmission of virus from HBV carrier-mothers to their children is the major mode of transmission in Taiwan [Stevens et al., 1975; Sung and Chen, 1978]. However, it was found that children from families with HBsAg-positive mothers or fathers had a comparable rate of HBsAg-positive (20% vs. 50%), suggesting paternal transmission through close contact may also be significant for intrafamilial HBV spread in Taiwan [Lin et al., 1991; Lin et al., 2005]. Similar findings of father to child transmission were demonstrated in other countries [Lin et al., 1991; Zhuo et al., 2000; Erol et al., 2003; Shimizu et al., 2003; Tajiri et al., 2007]. These results strongly indicate that father-to-child transmission is also a significant route of HBV infection.
Although HBV serological markers in the family contacts as shown in the family trees can reveal the possible routes of intrafamilial HBV transmission in most instances, identifying transmission routes in families with both parents positive for HBsAg is not possible with serological evidence alone. Comparison of nucleotide sequences from different HBV strains has been used to demonstrate the transmission of HBV in given populations [Dumpis et al., 2001; Zampino et al., 2002; Thakur et al., 2003; Tajiri et al., 2007]. A high homology between sequences isolated from different individuals strongly supports the viral strains originating from the same source. However, sequencing of viral genomes and subsequent homology comparison or phylogenetic analysis are tedious and labor intensive and thus cannot be applied to large samples. Genotyping of HBV by real-time PCR may be a useful tool for epidemiological investigation of intrafamilial HBV transmission. HBV genotypes were therefore identified in family members positive for HBsAg by using a real-time PCR based on the hybridization probes assay system, and the possible routes of intrafamilial transmission were determined (Fig. 1). Since HBV genotype B and C are the most prevalent genotypes in Taiwan [Kao et al., 2000; Kao and Chen, 2002], it is possible that children infected by intrafamilial members with the same genotype or extrafamilial horizontal transmission with homotypic but different strains. Therefore, to identify the routes of transmission, further sequence determination and phylogenetic analysis are needed.
In this study, four of 84 HBV vaccinees have intrafamilial HBV infections. The phenomenon of vaccine failure may be caused by maternal transmission, vaccine escape variants, and hypo- or non-response to the vaccine. Since the present study was cross-sectional rather than longitudinal, there were no previous data on the initial response of the children to vaccination. It is unclear whether they did not respond to vaccine and were infected, or they were infected by these vaccine escape mutants from the beginning of infection.
Previous studies from Taiwan, China, Japan, Hong Kong, Singapore, Thailand, India, Germany, the UK, the USA, Brazil, West Africa, and elsewhere had shown that various mutations and multiple variants within the “a” determinant (amino acid 124–147) and MHR (major hydrophilic region, amino acid 99–169) of HBsAg affect the antigenicity of protein which enables the virus to escape neutralizing antibodies [Carman et al., 1990; Karthigesu et al., 1994; Mimms, 1995; Oon et al., 1995; Oon et al., 1996; Lee et al., 1997; Nainan et al., 1997; Hsu et al., 1999, 2004, 2010; Zuckerman, 2000; Echevarría and Avellón, 2006; Ni et al., 2007]. The most frequent and stable mutation reported is the amino acid 145 variant, in which a single amino acid substitution of glycine to arginine (G145R) occurred at amino acid 145 of the “a” determinant of the surface antigen. A large study in Singapore of 345 infants born to mothers with HBsAg and HBeAg who received hepatitis B immunoglobulin at birth and plasma-derived hepatitis B vaccine at a dose of 5 µg or 10 µg within 24 hr of birth and then 1 month and 2 months later revealed 41 breakthrough infections with HBV, despite the presence of anti-HBs. The most frequent variant was G145R found in Singapore. Conversely, there was no evidence of infection among 670 immunised children born to mothers with HBsAg and anti-HBe, nor in any of 107 immunized infants born to mothers without HBsAg given the vaccine alone [Oon et al., 1995]. Another study in the USA showed that 94 (8.6%) of 1092 infants born to carrier mothers became HBsAg-positive after receiving postexposure prophylaxis with hepatitis B immunoglobulin and hepatitis B vaccine. Mutations in the “a” determinant of HBsAg were found in 22 children, most being in the 142–145 position—five had a mixture of wild-type HBV and variants, and 17 had only the 145 variant [Nainan et al., 1997]. HBV isolates carrying such mutations are believed to cause infection in infants and adults who have been vaccinated and/or received hepatitis B immunoglobulin. To investigate whether HBsAg seropositivity in these children was caused by mutations which alter the immunoreactivity of HBsAg, nested PCR and direct sequencing of S gene were undertaken. The most important and best-documented G145R mutation was not found, but other substitutions within the “a” determinant and MHR were found. Furthermore, one child (Fam2-Son) had a G145A mutation; but his mother was infected by glycine-145 (G145) wild-type HBV, suggesting that this mutant was selected and emerged following vaccination. The occurrence of these vaccine-escape mutants might threaten the ongoing HBV control strategies. Hsu et al. investigated whether vaccine escape variants affect vaccination efficacy [Hsu et al., 1999, 2004, 2010]. They demonstrated that the “a” determinant mutants had an advantage in infecting immunized children, and the prevalence of this mutant in HBV DNA positive children was 7.8% (8/103) in 1984 (just before vaccination was introduced) which increased significantly to 19.6% (10/51) in 1989, peaked at 28.1% (9/32) in 1994, and remained at 23.1% (3/13) in 1999 and 22.6% in 2004 [Hsu et al., 2010]. The results showed that there was no steady increase of the vaccine-escape mutants in vaccinees, and there was no evidence of the spread of these viruses, possibly because of weakness of the mutant [Kalinia et al., 2003].
Another possible cause of intrafamilial transmission in vaccine failure subjects is hypo- or non-response to the vaccine. Hypo- and non-responsiveness have been shown and associated with certain HLA types [Martinetti et al., 2000; Hsu et al., 1993]. Numerous amino acid differences occurred in the pre-S gene (showed in Fig. 5), these variations were within the immune epitope site. It might cause alterations of the immune target sites leading to escape from immune surveillance, or reduced binding affinity by MHC class I-mediated presentation of modified oligopeptides on the cell surface of hepatocytes [Milich et al., 1990]. A recent human trial had shown that a third-generation vaccine, containing pre-S antigen in addition to the S antigen, enhanced antibody responses in a population that included low responders and non-responders [Rendi-Wagner et al., 2006]. Thus, this new vaccine, incorporating pre-S1 and pre-S2 antigens, may help to overcome this problem and protect children from HBV infection.
In conclusion, this study demonstrated that introduction of universal HBV vaccination in infancy resulted in a decline in the overall HBsAg-positive rate and a reduction in intrafamilial transmission. Mother- or father-to-infant transmission remains the principal possible cause of vaccine failure that needs to be overcome. Furthermore, vaccine failure with respect to HBV variants and mutants as presented in this study is very important and should be considered a very important message. To prevent further HBV infection in the future, identifying subjects at a high risk for intrafamilial transmission and effective strategies or vaccines to block this transmission are required.




