Prediction and identification-based prediction of Chinese hepatitis C viral-specific cytotoxic T lymphocyte epitopes
Abstract
Cytotoxic T lymphocytes (CTLs) play a critical role in the host immune response to infection by the Hepatitis C Virus (HCV). In the current study, a number of HCV CTL epitopes that represent the HLA polymorphisms found in the majority of Chinese people were predicted based on genomic and bioinformatic approaches. The predicted epitopes were evaluated for validity by examining the peptide-binding affinity for MHC class I molecules, the stability of peptide–MHC complexes, and frequencies of IFN γ-positive T cells. Among the predicted epitope peptides, HLA-A2 restricted epitopes [NS4B (1793–1801) SMMAFSAAL] and HLA-B7 restricted epitopes [P7 (774–782) AAWYIKGRL] were able to induce high frequencies of IFN γ-producing T cells, and the specific CTLs for other epitopes were not detected in peripheral blood lymphocytes from patients with HCV. Moreover, NS4B (1793–1801) exhibited high binding affinity for HLA-A2 molecules, and its stability of peptide–MHC class I complexes was sufficient, indicating that the high binding affinity for MHC class I molecules is an important factor for immunogenicity. Primary analyses of the immunogenicity of predicted epitopes, such as in the current study, will contribute to the future design of an efficient vaccine that will be able to induce vigorous, sustainable, and broad HCV-specific CTL responses for the Chinese population. J. Med. Virol. 83:1315–1320, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Hepatitis C Virus (HCV) is the main pathogen responsible for chronic liver disease. Currently, 170–2,000 million people worldwide are infected with HCV, and the annual increase is estimated at about 3.5 million [Burra, 2009]. In China alone about 40 million people (3.2% of the population) are infected with HCV, and the subtypes -1b, -2a, and -3b, and type 2b predominate [Sun et al., 2009]. About 50–85% of all individuals infected with HCV develop chronic hepatitis, and 20–30% of those patients progress to liver cirrhosis that may lead to hepatocellular carcinoma (HCC) [Higuchi et al., 2002]. Despite HCV infection being a recognized threat to human health and characterized as a global health issue [Lavanchy, 2009], no ideal anti-virus treatment has thus far been established [Webster et al., 2009]. Clinical application of interferon- and Ribavirin-based therapy has proven suboptimal due to side effects and high cost [Bihl and Negro, 2009]. The high mutational rate of HCV and lack of effective in vitro or whole animal model systems have restricted the development of improved and novel HCV vaccines [Stoll-Keller et al., 2009].
T cell immune responses, especially the activities of the cytotoxic T lymphocytes (CTLs), are known to be responsible for the immune response against pathogens that have established intra-cellular residence [Chang, 2003]. Viral clearance in chronically infected HCV patients has been positively associated with increased CTL [Ishii and Koziel, 2008]. Thus, many studies have focused on characterizing the functions of HCV CTL cell epitopes, in hopes of exploiting their immune properties to treat and prevent HCV infection [Yerly et al., 2008]. Unfortunately, the heterogeneity and collection of quasispecies of HCV genomes continue to hinder the realization of this therapeutic goal [Söderholm et al., 2006]. To overcome these challenges, we sought to determine whether select specific epitopes that had been evolutionarily conserved among the different HCV isolates and their MHC limitation types could effectively promote immunogenicity.
MATERIALS AND METHODS
Bioinformatic Prediction of HCV CTL Epitopes
Database search keywords including HCV and genome polyprotein were used to extract relevant publications from the uniprot database (http://beta.uniprot.org/). Prevalent HCV genotypes (1b, 2a, 2b, 3, 3, 6a) in China were further used to search the protein database. The PepCleave algorithm (http//:peptibase.cs.biu.ac.il/PepCleave_II/) was adopted to predict potential proteasome hydrolysis nonapeptides and Bad Overweight Score as the substitution matrix. Compilation of a language script was used to handle the export sequence by PepCleave, to reserve only the nonapeptide portion, and save data in the FASTA format. One-hundred percent of homogenous sequences were identified and removed by use of Blast comparison.
The Hidden Markov Models method (HMM) of PREDTAP (http://antigen.i2r.a-star.edu.sg/predTAP) was used to calculate the binding capacity between the proteasome hydrolysis nonapeptides selected above and the transporter associated with antigen processing (TAP) protein; nonapeptides with high TAP-binding scores were retained. In consideration of the published findings [Yang et al., 2006], we adopted HLA alleles (In reference, A-B-DRB1 haplotypes were determined, and our study, HLA-A, HLA-B loci were determined) HLA A2: (A*0201, A*0205), A3: (A*3, A*1101, A*3302), A24: (A*24); B44, and B44: (B*40), which were most prevalent among the Chinese population. BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind) was used to forecast the binding conditions between candidate nonapeptides and HLA-1 A2, A24, A3, B7, B40 supertype molecules, respectively.
Possible CTL epitopes were evaluated, and the virus strains and genotypes covered by these CTL epitopes were predicted for the supertype molecules HLA-1A (A2, A3, A24) and B7 and B40 molecules. In order to comprehensively evaluate the scores of different CTL epitopes, the total score of CTL epitope antigenic peptide for HLA-1A or HLA-1B molecules was calculated by the function 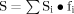 , where Si was equal to the corresponding TAP-binding scores between the HLA-1 supertype and predicted CTL epitopes, and fi equaled the phenotype frequency of this supertype in Chinese populations.
, where Si was equal to the corresponding TAP-binding scores between the HLA-1 supertype and predicted CTL epitopes, and fi equaled the phenotype frequency of this supertype in Chinese populations.
Synthesis of Polypeptide
The peptides were synthesized according to the amino acid sequence of the epitope selected above by means of a 431A polypeptide synthesizer and were purified to 98% homogeneity by reverse phase HPLC. Peptide aliquots were dissolved in 100% DMSO (Sigma-Aldrich, St. Louis, MO) at 4–20 mg/ml for binding. When peptides were used in cell culture, they were reconstituted at 20 mg/ml in DMSO and further diluted to 1 mg/ml with RPMI 1640 (Life Technologies, Carlsbad, CA).
Blood Samples
Fifty-six patients, aged 15–62 years old, with persistent HCV infection were admitted to Tangdu Hospital, Fourth Military Medical University between 2007 and 2009. Among them, 38 were men and 18 were women. All 56 patients were screened upon admission by class I HLA serotyping analysis, and were diagnosed with persistent HCV infection if the following criteria were met: HCV Ab detected by HCV ELISA test system, presence of HCV RNA by RT-PCR, elevated serum alanine aminotransferase (ALT) activity recorded for the past six months, and no known cause of chronic liver disease. The HCV genotypes of all patients were analyzed. Informed consent was obtained from all patients prior to participation in the study. The study protocol was approved by the local ethics committee and was carried out in conformity with the guidelines of the Helsinki declaration.
Peripheral blood mononuclear cells (PBMCs) from patients and normal healthy donors were separated on Ficoll–Histopaque density gradient (Sigma), washed three times in HBSS, and used for culture immediately or cryopreserved in media containing 80% fetal calf serum (Life Technologies), 10% DMSO (Sigma), and 10% RPMI 1640 (Life Technologies).
IFN γ Enzyme-Linked Immunosorbent Spot (ELISPOT) Assays
PBMCs from HCV infected patients and seronegative individuals were isolated and assayed by IFN γ ELISPOT as previously described [Tian et al., 2007]. The Invitrogen IFN γ ELISPOT kit (Carlsbad, CA) was used to detect human IFN γ, according to the manufacturer's instructions. The number of spots detected in assay wells with medium alone was subtracted from the number of spots in the experimental wells to determine IFN γ induction. PBMCs from HCV seronegative individuals were included in each experiment as negative controls. The cut-off for each patient was defined as twice the number of spots obtained with medium alone or as 50 spots per 106 cells when the background was less than 50 spots.
Cell-Based HLA Class I Binding Assay
T2 cells are human lymphoblastoid cells, and are MHC class II negative. This cell, in culture, also exhibits the characteristic defect in assembly and transport of MHC class I molecules. This defect results in a 70–80% reduction of HLA-A2 expression on the cell surface. HLA-A2 expression on T2 cells increases after exposure to peptides which can bind to HLA-A2. Therefore, this cell line is commonly employed in studies involving HLA-A2-restricted peptide epitopes [Wu et al., 2009].
The peptide-binding assay was performed as previously described [Sieker et al., 2009]. Briefly, T2 cells were suspended in AIM-V serum-free medium supplemented with 100 nM human beta-mercaptoethanol (β2m, Sigma), and were incubated with synthetic peptide at various concentrations at 37°C overnight. The next day, cells were stained for HLA-A2 expression by indirect immunofluorescence using the anti-HLA-A2 monoclonal antibody followed by FITC-labeled goat-anti-mouse IgG antibodies. Mean fluorescence intensities (MFI) were determined in the viable T2 cells according to the property of propidium iodide exclusion on a FACSort flow cytometer (Becton Dickinson, Shanghai, China). The concentration of each peptide that produced the half-maximal MFI of T2 cells pulsed with a control peptide was calculated as the half-maximal binding level (BL50). The known HLA-A2 epitope YLLPRRGPRL was used as a positive control peptide; the peptide LPGCSFSIF served as a negative control. For most peptides, experiments were performed three times, and data are given as mean ± standard error of the mean (SEM).
Complex Stability Assay
Peptides that showed a BL50 of less than 100 µM in the peptide-binding assay were further evaluated to determine their half lives as peptide–MHC class I complexes by means of the complex stability assay as previously described [Bui et al., 2006]. In brief, T2 cells were incubated with 100 µM peptide and 100 nM human β2m (Sigma) at 37°C overnight. Then, the cells were incubated for 1 hr at 37°C in R-10 containing brefeldin A (Sigma) at 10 µg/ml in order to block the egress of new MHC class I molecules. At the indicated time points, an aliquot was removed and stained with FITC-labeled anti-mouse IgG antibody. For each peptide, experiments were performed three times, and data are given as mean ± SEM.
CTL Generation and Cytotoxicity Assay
PBMCs were isolated from the peripheral blood of HLA-A2 adult healthy volunteers by Ficoll–Hypaque (Sigma) density gradient separation. PBMCs were then cultured in serum-free X-Vivo 15 medium (Lonza) in six-well plates at 37°C and 5% CO2. After 2 hr, nonadherent cells were removed by gentle washing with phosphate-buffered saline (PBS) solution. The nonadherent cells were used for positive selection of CD8 T cells by means of anti-CD8 isolation kit (Invitrogen, Beijing, China). Adherent cells were cultured in serum-free X-Vivo 15 medium in the presence of recombinant granulocyte macrophage colony stimulating factor (GM-CSF; PeproTech, Jiangsu Province, China) and interleukin-4 (IL-4; PeproTech) (100 ng/ml each) in six-well plates at 37°C and 5% CO2 for five days; a maturation cocktail containing 1000 U/ml IL-1β, IL-6, TNF-α and 1 µg/ml PGE2 (all from peprotech) was added to the immature DC for the final two days. The six predicted peptides and the confirmed HLA-A2 restricted peptide (Core (35–44) YLLPRRGPRL, as positive control) were pulsed on the mature DC. DC:CD8 T at the ratio of 5:1 were cultured in RPMI1640 with 10% AB serum, IL-7 (5 ng/m1), IL-12 (5 ng/ml), and IL-2 (5 ng/m1) in a 5% CO2 incubator at 37°C for seven days. Culture medium supplemented with the cytokines was refreshed every 48 hr.
The release of lactate dehydrogenase (LDH) was measured by using a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega). T2 cells were pulsed with the six predicted peptides and the positive peptides were used for target cells (1 × l05/ml cell density). T2 cells were plated into individual wells of a 96-well U-bottom plate in triplicate. Effector cells were added at an effector:target ratio (E:T) of 20:1, 10:1, or 5:1. The cell mixture was incubated for 4 hr at 37°C. Spontaneous release of target and effector cells was controlled by separate incubation of the respective populations; the cytotoxicity assay was evaluated according to manufacturer's instructions with the absorbance being measured at 490 nm on a microplate reader. The results were calculated as the mean of a triplicate assay. The percentage of specific lysis was calculated according to the following formula: cytotoxicity (%) = [(experimental − effector spontaneous − target spontaneous)/(target maximum − target spontaneous)] × 100%.
Statistical Analyses
Student's t-test was performed for all samples by using the SPSS v12.0 Software Package (Chicago, IL). Comparison among groups required multiple comparisons of multiple sample means. In these cases the LSD t-test was adopted, and the paired t-test was used to conduct intra-group comparisons and linear correlation analyses to analyze the correlation between the two data groups. Significant difference was considered having been reached when P < 0.05.
RESULTS
Bioinformatic Prediction of HCV CTL Epitopes
A protein database was constructed with the amino acid sequences of HCV genotypes 1b, 2a, 2b, 3a, 3b, and 6a, which were epidemic genotypes in China. The database included 17 proteome sequences, which were capable of forming a total of 51,799 nonapeptide monomers. This cohort consisted of 8,347 potential proteasome hydrolysis (Score > 0) nonapeptides predicted by Pep Cleave, and 4,075 non-repeated nonapeptides from different sites and different virus strains. After Blast comparison, those sequences found to share 100% homogeneity were removed; then, 3,789 potential proteasome hydrolysis nonapeptides were chosen for further study. The binding capacity of these 3,789 nonapeptides to TAP was calculated; 514 nonapeptides were found to have TAP-binding scores greater than or equal to six. Nonapeptides that scored greater than 1 in the binding-capacity test were selected. In the selection of candidate CTL epitopes, we considered the integrated scores of nonapeptides, virus genotypes covered, proportion of virus strains in the HCV database in this study, and the predicted binding-capacity between nonapeptide and HLA molecules. Through these criteria we finally selected 12 CTL epitopes (Table I).
| CTL epitopes | Score | HLA-1 | Virus genotypes | Proportion of virus strains | Position |
|---|---|---|---|---|---|
| FLARLIWWL | 4282.64142 | A2, A3, A24 | 1b | 6/8 | NS2–3 (838–846) |
| VLSDFKVWL | 1431.24927 | A2, A3, A24 | 6a | 2/2 | NS5A (1991–1999) |
| ALYDVIQKL | 303.26641 | A2, A3, A24 | 3a, 3b | 2/3 | NS5B (2604–2612) |
| ALYDVVSTL | 305.69439 | A2,A3, A24 | 1b | 7/8 | NS5B (2593–2601) |
| SMMAFSAAL | 103.39518 | A2, A3, A24 | 2a, 2b | 4/4 | NS4B (1793–1801) |
| ALYGVWPLL | 59.14324 | A2, A3, A24 | 1b | 5/8 | P7 (789–797) |
| AALENLVVL | 16.262 | B7, B40 | 1b | 5/8 | E2-P7 (746–754) |
| AAWYIKGRL | 16.262 | B7, B40 | 1b | 6/8 | P7 (774–782) |
| GAAVGSIGL | 5.942 | B7, B40 | 1b, 2a, 2b | 10/14 | NS4B (1836–1844) |
| AEQFKQKAL | 16.156 | B7, B40 | 1b | 7/8 | NS4B (1725–1733) |
| AYAARVPEL | 39.39681 | A24 | 2b | 2/2 | E1 (335–343) |
| GLSPAITKY | 26.37267 | A3 | 2a | 1/2 | E2 (708–716) |
Detection of T Cell Responses Specific for Predicted Restricted Epitopes During HCV Infection
To document the existence of cellular immune responses specific for predicted epitopes during natural infection, we quantified IFN-γ producing T cells by ELISPOT assays after in vitro stimulation of isolated human PBMCs. We selected 35 patients who expressed the HLA-A2 allele, 16 who expressed HLA-B7, 4 who expressed HLA-A24, and 2 who expressed HLA-A3 (HLA typing was performed by the Blood Center in Xi'an, China).
PBMC samples derived from the 35 HCV infected HLA-A2 expressing patients were analyzed using six predicted epitopes. Recalled cellular responses specific for NS4B (1793–1801) SMMAFSAAL were detected in 34 of these patients (97.14%), and the number of IFN-γ dots produced by these 34 patients' PBMCs was significantly higher than that in the control group (P < 0.05). Thirty-four of the patients were HCV-2a/2b genotype. The remaining single patient was HCV-1a, but did not exhibit specific cellular responses. However, after PBMCs were stimulated with the other five peptides, the number of IFN-γ dots produced by the original 35 patients' PBMCs was not significantly different from that of the control group (P > 0.05).
After PBMCs from the 16 HCV infected HLA-B7 expressing patients (with HCV-1b genotype) were stimulated with four peptides, with the exception of [P7(774–782)AAWYIKGRL], the number of IFN-γ dots produced by 16 patients' PBMC stimulated with other three peptides was not significantly different from that of the control group (P > 0.05). Moreover, epitopes of E1 (335–343) were not able to recall IFN-γ-producing cells in the four HCV patients expressing HLA-A24, and E2 (708–716) was likewise ineffective in the two HLA-A3 expressing HCV patients. These data demonstrate that HLA-A2 epitopes [NS4B (1793–1801) SMMAFSAAL] and HLA-B7 epitopes [P7 (774–782) AAWYIKGRL] can be naturally processed and that specific T cell responses can be induced during HCV infection.
Binding of HCV CTL Peptides to HLA Molecules
Six predicted HCV CTL peptides (Table I) were analyzed for their ability to bind to HLA-A2 molecules. The peptide-binding assay was used to show the binding affinity of a peptide for HLA-A2 molecules by measuring the stabilization of HLA-A2 on the TAP-deficient cell line T2 pulsed with each peptide. According to the BL50 values shown in Table II, the peptides examined were classified into two categories of response: (1) high binders displaying BL50 values less than 100 µM, and (2) low binders displaying BL50 values greater than 150 µM. These data indicated that most of the predicted epitopes were low binders. NS2-3(838–846), NS5A (1991–1999), RDRP (2604–2612), RDRP (2593–2601), and P7 (789–797) exhibited the lowest binding capacities for the HLA-A2 molecules. On the other hand, NS4B (1793–1801) presented an extremely high binding affinity for the HLA-A2 molecules.
| HCV peptide | Amino acid sequence | BL50 (µM) | Stability (h) |
|---|---|---|---|
| NS2–3(838–846) | FLARLIWWL | 136.1 ± 1.2 | 2.9 ± 0.5 |
| NS5A (1991–1999) | VLSDFKVWL | 149.0 ± 0.8 | 2.4 ± 0.6 |
| NS5B (2604–2612) | ALYDVIQKL | 137.2 ± 1.1 | 2.7 ± 1.3 |
| NS5B (2593–2601) | ALYDVVSTL | 155.9 ± 1.3 | 2.1 ± 0.8 |
| P7 (789–797) | ALYGVWPLL | 172.6 ± 1.9 | 2.5 ± 1.5 |
| NS4B (1793–1801) | SMMAFSAAL | 33.6 ± 0.7 | 17.5 ± 1.2 |
| Core(35–44) | YLLPRRGPRLa | 46.3 ± 1.6 | 13.3 ± 0.9 |
| Core(169–177) | LPGCSFSIFb | 132.8 ± 2.1 | 2.0 ± 1.3 |
- a The known HLA-A2 epitope was used as the positive control peptide.
- b The HLA-B7 peptide served as the negative control.
Half-Lives of Peptide–Class I Complexes
Six predicted HCV CTL peptides were further evaluated by the complex stability assay (Table II). We assumed that the long half-life of the peptide–MHC class I complexes would effectively increase the cell surface concentration of the peptide, thereby increasing the avidity of the cell–cell interaction. Thus, an epitope present at high cell surface concentration was expected to be able to better stimulate epitope-specific T cells. We found that in the cases of NS2-3(838–846), NS5A (1991–1999), RDRP (2604–2612), RDRP (2593–2601), and P7 (789–797) the half-life of the complexes was less than 3 hr. In contrast, the half-lives of complexes associated with NS4B (1793–1801) were longer than 8 hr. The data indicated that the stability of the peptide–MHC class I complexes was correlated with the binding affinity of a peptide for MHC class I molecules.
Peptide-Induced CTL Toxicity
CTL induced by peptide NS4B (1793–1801) SMMAFSAAL was able to lyse T2 cells that had been pulsed with peptide NS4B (1793–1801) SMMAFSAAL [(7.58 ± 0.16)%, (13.25 ± 0.62)%, (22.98 ± 0.96)% at ratios of 5:1, 10:1, and 20:1, respectively], which was significantly higher than that of the other five peptides (P < 0.05). This finding indicated that NS4B (1793–1801) was able to stimulate PBMCs from healthy donors to proliferate and induce antigen specific CTLs.
DISCUSSION
The objectives of this study were to identify HCV CTL antigen epitopes that are able to cover different types of HLA restriction corresponding to the majority of Chinese individuals and to provide a theoretical basis for the future study of antivirus HCV multi-epitope vaccines. In order to induce specific cellular immunity responses involving multiple HCV epitopes, some previous studies have pursued strategies based on logical combination of multiple epitopes selected from different parts of the HCV genome [Gao et al., 2006; Shi et al., 2006; Zeng et al., 2009]. An increasing amount of evidence has revealed that multiple epitopes from different parts of HCV genomes are necessary to generate HCV polypeptides with strong immunogenicity; moreover, these multi-epitope constructions have been found to be required to widely stimulate the host immune response mediated by the CD8+T cells [Mashiba et al., 2007]. The selection of coverage by HCV antigen epitopes plays a central role in the intensity and extent of induced cellular immunity response and the effectiveness of viral clearance [Stoll-Keller et al., 2009]. The immunogen is then subjected to a complicated processing mechanism in order to be presented by MHC and propagate the immune response. Presentation of MHC can activate the antigen epitopes of CD8+T cells [van der Burg et al., 2006]. Therefore, when designing multi-epitope vaccines, the selected epitopes must be able to cover the HLAs of different populations so that the vaccine will be effective across populations. However, most published HCV CTL epitopes were presented in HCV-infected patients from Europe and North America [Morita and Imawari, 2004; Wang et al., 2006; Matsueda et al., 2007], and it remains to be determined whether those epitopes are present in the Chinese.
We used the established molecular immunology of MHC-I presenting antigen (cytosol pathway) and relevant genomic knowledge, in conjunction with a bioinformatic analysis instrument, to generate a Chinese HCV protein database. We then employed multiple calculation methods to predict a set of potential CTL epitopes that covered most types of HLA restriction in the Chinese population.
The ideal prediction method of CTL epitopes reduced the large amount of candidate polypeptides down to a reasonable number [Li et al., 2008]. Therefore, the appraisal process was significantly simplified and appraisal accuracy enhanced. This will likely provide insight into the immune recognition mechanism of the CD8+T cell [Lundegard et al., 2006; Tong et al., 2007]. To document the existence of cellular immune responses specific for our predicted epitopes during natural infection, we quantified the IFN-γ producing T cells by ELISPOT assays after in vitro stimulation of patients' PBMCs. The results showed that HLA-A2 epitopes [NS4B (1793–1801) SMMAFSAAL] and HLA-B7 epitopes [P7 (774–782) AAWYIKGRL] were naturally processed and that specific T cell responses were induced during the HCV infection. The specific CTL for other epitopes was not detected in peripheral blood lymphocytes from patients with HCV.
In the current study, six predicted HLA-A2 restricted, HCV-derived peptides were evaluated for their peptide-binding affinity for MHC class I molecules and the stability of the resulting peptide–MHC complexes. Among the six predicted peptides [NS4B (1793–1801) SMMAFSAAL] bound to HLA-A2 with high affinity, the value of BD50 was 33.6 µM and the dissociation time of 50% HLA-A2 peptide complex was over 15 hr. Moreover, this predicted peptide could induce CTL reaction and CTL generation of PBMCs. Our results suggest that NS4B (1793–1801) might be an HLA-A2 restricted CTL epitope in the Chinese. This finding could provide the basis for future research on HLA-restricted polypeptide vaccines for Chinese people.
CONCLUSION
In summary, through this study we predicted several HCV CTL epitopes in Chinese individuals by genomic and bioinformatic analysis. Our results suggest that the HLA-A2 epitope [NS4B (1793–1801) SMMAFSAAL] and the HLA-B7 epitope [P7 (774–782) AAWYIKGRL] are potential HCV CTL epitopes in the Chinese. This information may contribute to the design of an effective HCV vaccine, which could induce vigorous and broad CTL responses specific for the Chinese population.
Acknowledgements
We would like to thank Xie Yuwei and Jin Boquan for their technical assistance.




