Diverse origin of P[19] rotaviruses in children with acute diarrhea in Taiwan: Detection of novel lineages of the G3, G5, and G9 VP7 genes†
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
Abstract
We previously reported the detection of genotype P[19] rotavirus strains from children hospitalized with acute dehydrating diarrhea during a 5-year surveillance period in Taiwan. The characterization of five P[19] strains (0.4% of all typed), including three G3P[19], a novel G5P[19], and a unique G9P[19] genotype is described in this study. Phylogenetic analysis of the VP4, VP7, VP6, and NSP4 genes was performed, which demonstrated novel lineages for respective genotypes of the VP4 and the VP7 genes. The sequence similarities of the P[19] VP4 gene among Taiwanese human strains was higher (nt, 91.5–96.2%; aa, 93.7–97.6%) than to other P[19] strains (nt, 83.5–86.6%; aa, 89.4–94.1%) from different regions of the world. The VP7 gene of the three G3P[19] Taiwanese strains shared up to 93.4% nt and 97.5% aa identity to each other but had lower similarity to reference strain sequences available in GenBank (nt, <90.1%; aa, <95.6%). Similarly, the VP7 gene of the novel G5P[19] strain was only moderately related to the VP7 gene of reference G5 strains (nt, 82.2–87.3%; aa, 87.0–93.1%), while the VP7 gene of the single G9P[19] strain was genetically distinct from other known human and animal G9 rotavirus strains (nt, ≤92.0%; aa, ≤95.7%). Together, these findings suggest that the Taiwanese P[19] strains originated by independent interspecies transmission events. Synchronized surveillance of human and animal rotaviruses in Taiwan should identify possible hosts of these uncommon human rotavirus strains. J. Med. Virol. 83:1279–1287, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Group A rotaviruses are the leading cause of severe dehydrating gastroenteritis in young children worldwide. Globally, rotavirus infections account for an estimated 611,000 deaths each year among children <5 years of age [Parashar et al., 2006]. Rotavirus particles are icosahedral, 70–75 nm in diameter and contain 11 genomic segments of double-stranded RNA. The two outer layer proteins VP7 and VP4 form the basis of the dual classification system of group A rotavirus into G and P types [Estes and Kapikian, 2007]. Of the 24 G and 31 P genotypes currently known, at least 11 G types and 12 P types, in at least 70 different antigen combinations have been identified in humans [Matthijnssens et al., 2009a]. Of these, five are globally common (G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8]), while others are unusual combinations of these common antigen types and may be only regionally important. Rare strains include mainly those antigen specificities that originate from periodic occurrence of animal-to-human interspecies transmission events. Examples for such strains include G5, G6, G8, and G11 VP7 genes or P[3], P[7], P[11], and P[19] VP4 genes, as well as rare genetic variants of common antigen types, such as G3 VP7 genes of porcine, feline, or lapine origin or genetically diverse porcine-like P[6] or feline-like P[9] VP4 genes [Bányai et al., 2003, 2009a,b,c; Khamrin et al., 2006; Martella et al., 2006, 2008; Matthijnssens et al., 2006, 2009b; Steyer et al., 2008; Chitambar et al., 2009; Esona et al., 2009; De Grazia et al., 2010].
Based on sequence similarities, recently a new classification system has been proposed, which designates independent genotypes for each genome segment [Matthijnssens et al., 2008a,b]. This system is being appreciably adopted by the scientific community and several research groups initiated partial or whole genome-based strain characterization studies to extend information on features of circulating rotavirus strains. For example, among the nine non-serotype genes, the VP6 and NSP4 genes have been studied extensively due to their recognized relative genetic linkage and their potential use in determining the original host species for unusual strains with suspected heterologous origin [Iturriza-Gomara et al., 2003; Khamrin et al., 2006, 2007; Araújo et al., 2007; De Grazia et al., 2007, 2008, 2009; Ghosh et al., 2007; Li et al., 2008; Martella et al., 2008; Bányai et al., 2009; Chitambar et al., 2009; Khamrin et al., 2009].
In Taiwan, rotavirus strain surveillance started in 2001 [Chen et al., 2005]. Studies have identified the globally common strains, although the predominant antigen types seem to change over time. We have recently reported strain prevalence data over a 3-year period from three sentinel hospitals located in northern, central, and southern Taiwan [Wu et al., 2009]. That study utilized a nucleotide sequence-based genotyping assay, in which type common PCR primers were employed to amplify full-length or partial outer capsid genes. In total 829 strains were genotyped. Among these strains we identified G1P[8] strains to be predominant, followed by G3P[8], G9P[8], and G2P[4] strains as additional medically important strains, while 11 further antigen combinations represented minority strains. Unexpectedly, among these minority combinations we detected two P[19] strains associated with a G3 and a G9 VP7 gene. In subsequent extended surveillance, we detected three additional P[19] strains in 2008 and 2009 seasons; two associated with G3 and one with G5 VP7 gene. In an attempt to better understand the origin and relationship of these strains to one another and the few hitherto characterized human and animal P[19] strains, we determined and analyzed the full-length or nearly full-length VP7 genes, the variable VP8* fragment of the VP4 gene, and the genotype diversity and potential genetic linkage among the NSP4 and the VP6 genes.
MATERIALS AND METHODS
Case Definition and Data Collection
From January 2005 to December 2009, a total of 6,346 stool specimens were collected from children <5 years old who were hospitalized for treatment of acute gastroenteritis (AGE) in three sentinel hospitals located in northern, central, and southern Taiwan. AGE was defined as three or more episodes of watery diarrhea or looser than normal stool in the 24 hr before presentation.
Laboratory Testing
Stool specimens were collected and screened for rotavirus VP6 antigen using an EIA kit (RIDASCREEN® Rotavirus, R-Biopharm AG, Darmstadt, Germany) at each hospital. Rotavirus EIA-positive specimens were shipped to Rotavirus Reference Laboratory at Taiwan CDC for G- and P-genotyping.
RT-PCR and Polyacrylamide Gel Electrophoresis
Viral RNA was extracted from 10% (w/v) fecal supernatants by MagNA Pure LC DNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. The extracted RNAs were used as the template for RT-PCR with random primers [Wu et al., 2009]. Viral VP4, VP6, VP7, and NSP4 genes were amplified with primers sets Con3/Con2, JRG7/JRG8 or GEN-VP6F/GEN-VP6R, Beg9-End9, and JRG30/JRG31 as described [Gouvea et al., 1990; Gentsch et al., 1992; Matthijnssens et al., 2006; Esona et al., 2009; Luan le et al., 2009]. Viral RNA samples were also analyzed by a 10% polyacrylamide gel and silver staining according to the manufacturer's protocol (Bio-Rad, Hercules, CA).
Sequencing and Phylogenetic Analysis
PCR amplicons were directly sequenced with primers sets Con3/Con2, JRG7/JRG8 or GEN-VP6F/GEN-VP6R, Beg9-End9, and JRG30/JRG31 on a ABI 3130 sequencer (Applied Biosystems, Foster City, CA). Rotavirus genotypes were determined using the RotaC software [Maes et al., 2009]. Multiple nucleotide sequences were aligned and the phylogenetic analysis was performed using MEGA 4.0 software [Tamura et al., 2007].
RESULTS
Genetic Analysis of the VP4 Genes
A 831 bp fragment of the VP4 gene (corresponding to nt 34–864, strain Mc345) of five Taiwanese P[19] strains was sequenced. Along a shorter stretch (750 bp and 250 aa), these strains shared 91.5–96.2% (nt) and 93.7–97.6% (aa) identities among each other and a lower similarity to 16 other P[19] strains detected in mainland Asia (range, nt, 83.5–86.6%; aa, 89.4–94.1%). In our phylogenetic analysis, both nt (Fig. 1A) and aa (not shown) trees demonstrated that Taiwanese P[19] strains fell into a discrete genetic lineage. An aa alignment of the VP8* fragment revealed 5 aa substitutions in the VP8* fragment of the protein, which were unique for the Taiwanese P[19] strains (Fig. 1B).
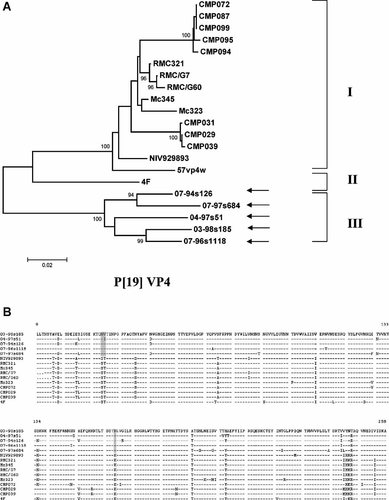
A: Nucleotide sequence-based phylogenetic trees of the P[19] VP4 gene. The arrows indicate the Taiwanese G3P[19], G5P[19], and G9P[19] strains. Bootstrap values >90% are indicated. Scale bars are proportional to the phylogenetic distance. Proposed lineage designation of the P[19] VP4 gene is indicated with I, II, and III, respectively. B: Amino acid alignment of the VP8* domain of VP4 gene. Lineage specific substitutions characteristic to Taiwanese strains are highlighted. Identical residues are shown with dashed line.
Genetic Analysis of the VP7 Genes
As determined by nucleotide sequence-based genotyping, the five Taiwanese P[19] strains exhibited three different VP7 specificities: G3 (03-98s185, 07-94s126, 07-97s684), G5 (04-97s51), and G9 (07-96s1118) (Fig. 2).
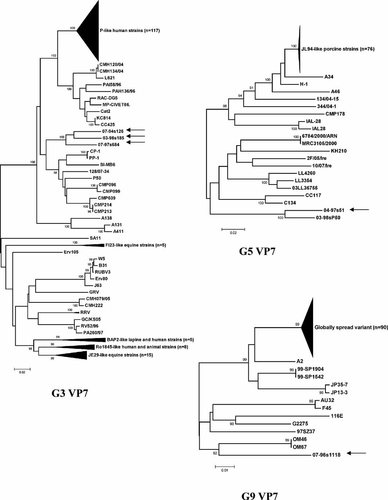
Nucleotide sequence-based phylogenetic trees of the G3, G5, and G9 VP7 genes. The arrows indicate the Taiwanese G3P[19], G5P[19], and G9P[19] strains. Strain designated 03-98sP50 is a G5P[6] Taiwanese strain. Bootstrap values >90% are indicated. Scale bars are proportional to the phylogenetic distance.
The full or nearly full-length coding region was determined for the G3 strains. These three strains shared 92.8–93.4% nt (and 96.3–97.5% aa) similarities along a 972 bp long fragment to each other, and a lesser similarity to 183 reference G3 strains (nt, <90.1%; aa, <95.6%). They are most closely related to porcine isolates (e.g., CP-1). Along the antigenic region these three Taiwanese strains shared high similarities, although 1–4 aa substitutions were seen (Fig. 3). Phylogenetic analysis found that these three strains clustered on a separate branch, distinct from other strains suggesting they represent a new lineage of the G3 VP7 gene (Fig. 2).

Primary structure of the predicted VP7 antigenic sites A, B, C, and F of Taiwanese and reference G3, G5, and G9 rotavirus strains. Dashed line indicates amino acid residue identities in the alignment.
Identification of a G5P[19] strain in our sample collection was unexpected, as this antigen combination was not identified previously in humans. Sequence analysis of a 828 bp stretch of the VP7 gene of strain 04-97s51 revealed moderate sequence identity to 98 reference G5 strains of both animal and human origin (range: nt, 82.2–87.3%; aa, 87.0–93.1%). Similar divergence was seen in the neutralization epitopes between 04-97s51 and selected G5 strains. Overall, these similarity ranges together with results of phylogenetic analysis suggested that this Taiwanese G5 strain fell into a new lineage of the G5 VP7 gene (Fig. 2).
A fifth Taiwanese P[19] strain contained a G9 VP7 gene. Because this specificity was seen in both human and porcine strains in South-East Asia, it was interesting to see whether the Taiwanese isolate clustered with these human or animal rotavirus strains. Of note, the 07-96s1118 strain displayed limited sequence similarity with other G9 strains detected in Taiwan or other parts of the world. Overall, this Taiwanese G9 strain shared ≤92.0% nt (and ≤95.7% aa) identity to any known G9 VP7 gene along the full coding region, including both human and porcine strains, and shared particularly lower similarity to the globally emerging variant of G9 VP7 gene (nt, <90%). The closest relatives were among the unique US OM46-like G9 strains that were detected during 1997–1998 in Omaha (nt, 92%; aa, 95.1%). The neutralization epitopes of this novel Taiwanese G9 strain also suggested extensive divergence from other G9 strains. All these data demonstrated that 07-96s1118 is not only highly divergent from the globally emerging variant of G9 VP7 gene, but also divergent from all other G9 VP7 genes identified in surveillance studies over the last 30 years (Figs. 2 and 3).
Genetic Analysis of the VP6 and NSP4 Genes
The full or partial coding regions of the VP6 and NSP4 genes were determined to further characterize the five identified Taiwanese P[19] strains. For the VP6 gene, the RotaC [Maes et al., 2009] online genotyping software that was developed to predict rotavirus A genotype specificities identified two VP6 genotypes among Taiwanese P[19] strains (Fig. 4). Four strains were classified as genotype I1, a common human VP6 specificity, while 07-96s1118, the unique Taiwanese G9P[19] strain shared only moderate sequence identity to other genotypes. The automated genotyping platform recommended validating the VP6 gene specificity of this strain through the RCWG, whose opinion was that this gene falls within the newly described I12 VP6 genotype. This genotype is currently represented by two strains, the reference human Bangladeshi G11P[25] strain, KTM368, and the Taiwanese G9P[19] strain, 07-96s1118. Phylogenetic analysis suggested this genotype is intermediate between genotype I1 (≤85%) and I5 VP6 (≤86%) genotypes and even its representative strains share only marginal nt identities to one another (87.6% nt). The other four P[19] strains shared ∼90–98% nt identities among each other, and were most closely related to two porcine strains (i.e., 04-97s51-like P[19] strains vs. CN86, 97.8–97.9% nt identities and 03-98s185 vs. JL94, 91.1% nt identity).

Nucleotide sequence-based phylogenetic trees of the VP6 and the NSP4 genes of Taiwanese P[19] strains (the arrows indicate these strains). Bootstrap values >80% are indicated. Scale bars are proportional to the phylogenetic distance.
RotaC clustered all five Taiwanese P[19] strains into a single E1 NSP4 genotype with ≤93.4% identity to reference strains (Fig. 4). The P[19] strains shared 93.0–96.0% nt similarities among one another. Among the reference strains, the rare human–porcine reassortant strains (e.g., LL4260) were most closely related to these Taiwanese P[19] strains (range: nt, 91.7–93.4%).
Three of the five Taiwanese P[19] rotavirus strains with adequate amount of RNA were further examined for their RNA profiles by polyacrylamide gel electrophoresis (Fig. 5). The two strains with G5P[19] and G9P[19] genotypes had long RNA electropherotypes similar to that of the reference Wa strain, whereas one G3P[19] strain, 03-98s185, had a super short electropherotype as indicated by its rearranged gene segment 11.
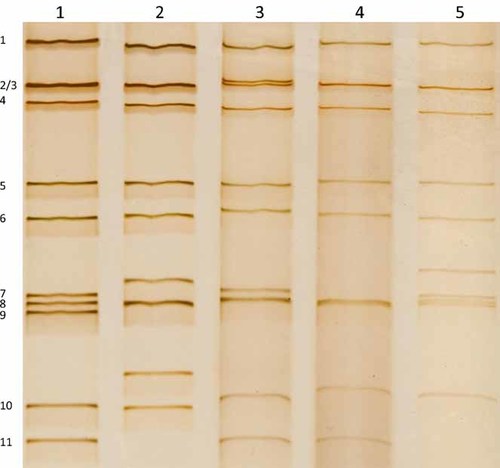
RNA profiles of P[19] rotavirus strains from children with diarrhea in Taiwan. Viral RNA was analyzed by polyactylamide gel electrophoresis and silver staining. Lanes 1–5, strains Wa, DS-1. 04-97s51, 07-96s1118, and 03-98s185, respectively. Segments of the genome from the reference strain Wa are shown on the left.
DISCUSSION
This study reports new findings of unusual P[19] rotavirus strains from our sentinel hospital-based surveillance during 2004–2009. Among 1,183 strains G and P typed, there were 5 isolates (0.4% of total) belonging to genotype P[19]. These isolates were identified from specimens collected from different years, one in 2005 (0.8%), one in 2007 (0.3%), two in 2008 (1%), and one in 2009 (0.6%). Thus, these rare strains were circulating among humans or alternatively in an animal host in Taiwan during most of the surveillance period.
There have been a few reports on rotavirus P[19] strain detection among animals and humans since 1990, beginning with the first report of the isolation and characterization of P[19] prototype strain 4F in 1994 [Burke et al., 1994]. Several other studies document detection of the virus in humans from Thailand [Urasawa et al., 1992], India [Varghese et al., 2004], and Vietnam [Nguyen et al., 2008]. In the present study, phylogenetic analysis revealed that the Taiwanese strains were genetically closely related to each other but were distinct from human or animal strains from other countries. For example, in the VP8* fragment of VP4 there were five conserved aa substitutions found in Taiwanese P[19] strains compared to human or animal P[19] strains from other countries. In addition, four other aa sites, positions 15, 44, 73, and 254, are more closely related to the prototype 4F strain than human or animal P[19] strains isolated from other Asian countries.
The most prevalent VP7 genotypes in Taiwan were G1 (48.3%), G3 (24.5%), G9 (14.1%), G2 (8.0%), and some rare genotypes like G4, G5, and G8 [Wu et al., 2009]. In most studies globally, G1 represented approximately 50–65% of the strains, followed by G3, G4, and G2 [Gentsch et al., 2005; Santos and Hoshino, 2005]. In addition, an increasing number of rare genotypes have been described recently, many of which have strong genetic relationships with animal rotaviruses [Matthijnssens et al., 2009b].
Among the G3 strains, we found three with P[19] specificity, one each from 2005, 2008, and 2009. The nucleotide sequences of these three strains showed only moderate similarity between them (only 92.8–93.4%), and less similarity to 183 reference G3 strains. The most closely related strain was a porcine isolate, CP-1 which clustered on a distinct branch by phylogenetic analysis. Demonstration that there were several amino acid substitutions in the antigenic sites of these Taiwan G3 strains supports the proposal that they represent a new G3 lineage.
Human G5 rotaviruses were first reported in combination with genotype P[8] as a regionally common strain in Brazil during 2004 [Alfieri et al., 1996]. Unusual human G5P[6] rotaviruses were identified from AGE children under five in China and Vietnam [Ahmed et al., 2007; Li et al., 2008] and more recently, a G5P[7] strain new to humans, has been detected in Cameroon [Esona et al., 2009]. However, G5 strains had not previously been reported in Taiwan. In addition, this is the first report on detection and molecular characterization of a G5P[19] strain. Molecular features of the G5 VP7 gene of our Taiwanese strain belonged to a phylogenetic lineage distinct from those represented by other strains.
Unlike the G5P[19] strain, the single Taiwanese G9P[19] strain is not the first representative of this particular antigen combination [Okada et al., 2000; Das et al., 2004; Mukherjee et al., 2010], however, it carried a G9 VP7 gene highly divergent from other currently known G9 VP7 genes, indicating that introduction of this particular gene into the human population in Taiwan was an independent evolutionary event from those events that occurred in other parts of the world during the past three decades. In a historical perspective, the DC706-like G9P[8] strains detected exclusively on the eastern coast of the USA represents the first documented evidence for human infection with G9 strains and could have caused a minor local outbreak [Cao et al., 2008]. Independent introduction associated with a different G9 VP7 gene lineage (WI61-like G9P[8]) was seen northeast from the site where DC706-like strains were detected during 1983 and this strain seemed to have some capacity to spread into other areas as evidenced by detections of closely related strains in subsequent years in Japan [Clark et al., 1987; Green et al., 1989]. A novel strain, Mc345-like G9P[19], was identified in Thailand in 1989 and its descendants were detected subsequently in this region [Urasawa et al., 1992; Okada et al., 2000; Das et al., 2004; Mukherjee et al., 2010]. Another novel strain, 116E-like G9P[11], appeared during early 1990s in India in asymptomatic neonates [Das et al., 1993; Gentsch et al., 1993]. The first description of the globally spread variant of G9 strains was also first detected in South Asia and genetically it was most closely related to Mc345-like strains. This variant of G9 strains that subsequently spread worldwide to become the 5th most common strain during a 10-year period are additional examples of introductions of G9 VP7 into human populations [Gentsch et al., 2005; Matthijnssens et al., 2009a]. Additional emergences seen in central region of the USA during mid-1990s by a divergent G9P[8] strain (OM46-like) [Laird et al., 2003], and other G9 strains (97'SZ37 and T203) with divergent VP7 genes were reported in far-east Asia [Xu et al., 2000, GenBank record; Dong et al., 2009]. While to date only the lineage III G9 strains have been proven to possess significant epidemiologic potential, the data on a new G9 VP7 gene variant from Taiwan clearly demonstrates that interspecies transmission events are more frequent and the G9 VP7 gene is genetically more divergent than previously thought based on a limited number of gene sequences.
Overall, our findings demonstrate that the P[19] strains circulating in Taiwan are different from those in other Asian countries. The distinct evolutionary patterns of these strains may be explained by Taiwan's geographic separation from the rest of Asia by the Pacific Ocean and consequently minimal genetic contact with rotavirus strains of suspect animal reservoirs from other countries. Since P[19] strains may have originated in porcine species, it will be important to conduct surveillance and determine whether similar strains are also present among pigs in Taiwan.
Acknowledgements
We thank Chen-Fu Yang for discussion and Shy-Yuan Liang for assistance in performing data management and molecular analysis, Yuhuan Wang for performing viral RNA analysis, and Jelle Matthijnssens for helpful discussion.




