Occult hepatitis C virus infection in Iranian patients with cryptogenic liver disease
Abstract
The diagnosis of cryptogenic liver disease is made when after extensive evaluations, recognizable etiologies of chronic liver disease are excluded. In this study, the presence of hepatitis C virus (HCV) RNA was tested in peripheral blood mononuclear cells (PBMCs) taken from Iranian patients who although were found negative for plasma HCV RNA and anti-HCV antibodies, suffered from chronic liver disease of unknown etiology. From September 2007 to March 2010, 69 patients from Tehran with chronic liver disease of unknown etiology who were referred to our center were enrolled in the present study. PBMCs were isolated from 10 mL peripheral blood specimens. HCV-RNA status was tested in plasma and PBMCs samples by reverse-transcription polymerase chain reaction (RT-PCR). HCV-RNA was detected in HCV-positive PBMCs specimens by RT-PCR method. HCV genotypes were subsequently analyzed in HCV-positive samples using restriction fragment length polymorphism (RFLP) assay; then HCV genotypes were confirmed by sequencing of 5′ non-coding fragments after cloning PCR products into pJET1.2/blunt cloning vector. HCV-RNA was detected in PBMCs specimens belonging to 7 (10%) out of 69 patients. Genotyping of the HCV-RNA isolated from PBMCs showed that 3 (43%) patients with occult HCV infection had genotype 1b, 2 (29%) had genotype 1a, and another 2 (29%) had genotype 3a. The results of this study suggest that patients with chronic liver disease of unknown etiology may have occult HCV infection in the absence of anti-HCV antibodies and plasma HCV-RNA. It has been suggested that in the absence of liver biopsy specimens, analysis of PBMC sample for HCV-RNA would be informative. J. Med. Virol. 83:989–995, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
In approximately 5% of patients with chronic liver disease, no etiology can be identified; such cases are known as cryptogenic liver disease. The diagnosis is made when after extensive evaluations recognizable etiologies of chronic liver disease such as viral hepatitis, autoimmune hepatitis, non-alcoholic steatohepatitis (NASH), metabolic liver disease, cytomegalovirus or Epstein-Barr virus infections, hepatotoxic drug, Wilson's disease, biliary tract disease, venous outflow obstruction, α1-antitrypsin deficiency, decompensated diabetes, thyroid dysfunction, alcohol abuse, iron overload, morbid obesity, or other systemic diseases, are excluded. In reviewing of studies from Iran, hepatitis B virus (HBV) infection was found as the most common cause of cryptogenic liver disease [Honarkar et al., 2005; Hollinger et al., 2010].
Hepatitis C virus (HCV) is a single-stranded RNA virus of the family Flaviviridae and genus Hepacivirus. This virus infects over 170 million individuals worldwide and causes chronic hepatitis in up to 85% of cases. In more than 20% of patients, chronic hepatitis C progresses to hepatic fibrosis and cirrhosis, and in approximately 3%–5% of cases eventually would result in hepatocellular carcinoma (HCC) [Lauer and Walker, 2001; Freeman et al., 2003; Seeff et al., 2003; El-Serag and Rudolph, 2007].
The prevalence rate of HCV in Iran is low; the rate in general population is less than 0.2% [Alavian et al., 2009]. Diagnosis of chronic HCV infection is based on the detection of specific anti-HCV antibodies and/or genomic HCV-RNA in serum for more than 6 months after acute infection [Castillo et al., 2004; Carreño, 2006].
Recently, a new form of chronic HCV infection was described by Castillo et al. [2004]. This infection is characterized by the absence of HCV-RNA and anti-HCV antibodies in serum in the presence of genomic HCV-RNA in the liver biopsy specimen, in patients who suffer from chronic liver disease of unknown etiology [Castillo et al., 2004]. Furthermore, about 70% of patients with occult HCV infection also have HCV-RNA in their peripheral blood mononuclear cells (PBMCs); the genomic and the antigenomic HCV-RNA have also been detected in these cells. The detection of antigenomic HCV-RNA strand in PBMCs of subjects with occult HCV infection can be supported the hypothesis that HCV is able to replicate in these cells. On the other hand, the antigenomic HCV-RNA strand has been detected in liver biopsy sample in 84% of patients with occult HCV infection, indicating that HCV is replicating in these cells [Castillo et al., 2004; Laskus et al., 2007].
Although detection of genomic HCV-RNA strand in liver biopsy specimen is the gold standard and the most accurate method for the diagnosis of occult HCV infection, testing for HCV-RNA in PBMCs is an alternative and easy to do when a liver biopsy is not available [Bartolome et al., 2007; Barril et al., 2008]. The objective of this study was to investigate the presence of occult HCV infection in Iranian patients with cryptogenic liver disease.
PATIENTS AND METHODS
Patients
In this cross-sectional study, 69 consecutive patients with established chronic liver disease of unknown etiology who were referred from September 2007 to March 2010 to hospitals affiliated to Iran, Baqiyatallah, Tehran, and Shaheed Beheshti University of Medical Sciences referral centers for liver diseases were studied. Twenty-three of 69 patients had previously undergone a liver biopsy for the diagnostic purposes and their formalin fixed paraffin-embedded liver biopsy specimens were available; the rest of the patients had been diagnosed by ultrasonography, clinical features, and laboratory findings. Fifty-six (81%) patients suffered from cryptogenic cirrhosis. Informed consent was obtained from the participants. The study was approved by the local ethics committee of Gastrointestinal and Liver Disease Research Center (GILDRC) of Tehran University of Medical Sciences, Tehran, Iran.
The inclusion criteria included (i) having sustained abnormal liver function tests with no known etiology for more than 6 months (tested every 3 months) before entry into the study; (ii) all of the patients were anti-HCV- (ETI-AB-HCVK-4, Diasorin, Spain) and serum HCV-RNA-negative (patients have been tested for anti-HCV and serum HCV-RNA more than two times before entry to the study); (iii) exclusion of all other known causes of liver disease based on clinical, laboratory, and epidemiological data, including infection by HBV (hepatitis B surface antigen and serum HBV-DNA negative), autoimmunity (negativity for antinuclear and antimitochondrial antibodies, etc.), genetic disorders, drug toxicity, alcohol intake, etc.; and (iv) persistently negative for anti-human immunodeficiency virus antibodies (anti-HIV Abs).
Collection and Preparation of Samples
About 10 mL of peripheral blood was collected from each patient into EDTA-containing vacutainer tubes. Plasma was separated from whole blood and stored at −70°C. PBMCs were isolated from 10 mL EDTA-treated blood by a standard procedure of Ficoll–Hypaque gradient centrifugation (Lymphoprep, Oslo, Norway). The PBMCs pellet was then washed three times with phosphate-buffered saline (pH = 7.4), counted and resuspended in 200 µL RNALater (Ambion Inc., Austin, TX, USA), and afterwards frozen at −70°C for later detection.
Plasma and PBMCs specimens taken from 25 patients with chronic HCV infection, as well as 25 samples taken from healthy blood donors were used as positive and negative controls, respectively.
Isolation of RNA From Plasma and PBMC Specimens
RNA was extracted from 140 µL of plasma using the QIAamp viral RNA extraction kit (Qiagen GmbH, Hilden, Germany), and from a pellet of approximately 3–5 × 106 PBMCs using the RNeasy minikit (Qiagen GmbH), according to the manufacturer's instructions. The amount of total RNA extracted from PBMC was determined by spectrophotometry. The total RNA extracted from plasma or 0.5 µg of RNA extracted from PBMC was used for genomic HCV-RNA detection.
HCV-RNA Detection in Plasma and PBMC Samples by RT-PCR
Detection of HCV-RNA in serum and PBMC specimens was performed by reverse-transcription polymerase chain reaction (RT-PCR) method. Briefly, cDNA synthesis of HCV-RNA was carried out at 42°C for 30 min in a 20-µL reaction mixture containing 0.5 µg of total RNA, 20 pmol of random hexamer, 4 µL of 5× reverse transcriptase reaction buffer, 125 µmol mix deoxynucleotide triphosphat, 104 U of Moloney Murine Leukemia Virus reverse transcriptase (Fermentas GmbH, St. Leon-Rot, Germany), 19.2 U of RNase inhibitor (Fermentas GmbH), and 1 µL of diethyl-pyrocarbonate (DEPC) treated water, followed by heating at 72°C for 10 min for inactivation of reverse transcriptase.
A set of nested primers from the 5′ non-coding region was used, including an outer primer pair of 244-base span: forward primer fap (5′-GCAGAAAGCGTCTAGCCATGG-3′; 68–88), numbering of nucleotide sequences as previously described [Choo et al., 1991], and reverse primer zap (5′-CTCGCAAGCACCCTATCAGGC-3′; 312–291) and an inner primer pair of 211-base span: forward primer fip (5′-TCTAGCCATGGCGTTAGTA-3′; 80–97), and reverse primer zip (5′-CAGTACCACAAGGCCTTTC-3′; 291–270) [Schröter et al., 2001, 2003].
The first round PCR was performed in a 50-µL mixture containing the template (5 µL of reverse transcription solution), 2.5 U of Taq DNA polymerase, 5 µL of 10× PCR buffer, 20 pmol of each outer primer, 100 µmol mix deoxynucleotide triphosphat, and 0.15 mmol MgCl2. Amplification was performed as follows: Initial denaturing for 4 min at 94°C and 30 cycles of 94°C for 30 sec, 60°C for 35 sec, and 72°C for 40 sec, followed by a final extension at 72°C for 4 min. After the first amplification, 5 µL of PCR product was added to a second PCR amplification with the inner primer pair. For nested PCR, the same procedure was performed as described for the first PCR. Amplification was performed as follows: Initial denaturing for 4 min at 94°C and 30 cycles of 94°C for 20 sec, 52°C for 20 sec, and 72°C for 30 sec, followed by a final extension at 72°C for 4 min. PCR products (214-bp) of specimens with positive and negative controls and DNA marker were visualized by 2% agarose gel electrophoresis. All the necessary precautions (guidelines of Kwok and Higuchi) were used to prevent contamination [Kwok and Higuchi, 1989], and appropriate controls (positive and negative serum controls) were included with each assay.
To determine the limit of sensitivity of RT-nested PCR, this method was tested by serial dilution of a plasma sample with known viral load (105 HCV-RNA copies/mL was determined by Cobas TaqMan 48 Analyzer [Roche Diagnotic GmbH, Mannheim, Germany]). The sensitivity of this study's method was 50 IU/mL plasma.
HCV-Genotyping and Sequencing of the 5′ Non-Coding Region
Hepatitis C virus-RNA extraction and reverse transcription were performed as described earlier. HCV genotypes were analyzed in HCV-positive PBMCs specimens using restriction fragment length polymorphism (RFLP) assay of the 5′ non-coding region of the HCV genome as described previously [Pohjanpelto et al., 1996].
Hepatitis C virus-genotyping by RFLP assay has also been confirmed by sequencing of the 5′ non-coding region fragments after cloning of the PCR products into pJET1.2/blunt cloning vector (Fermentas GmbH). Briefly, the 5′ non-coding region of HCV was amplified from total RNA isolated from HCV-positive specimens by RT-PCR assay as described above, but 3.5 U of pfu DNA polymerase were used instead of Taq DNA polymerase. The 214-bp PCR products of 5′ non-coding region were purified using high pure PCR product purification kit (Roche Diagnostic GmbH, Mannheim, Germany). The fragments were then cloned into the pJET1.2/blunt cloning vector, and three clones from each sample were sequenced by dye termination method using the ABI 3730 XL sequencer.
Phylogenic Analysis
The sequences were aligned with 5′ non-coding region sequences corresponding to all HCV genotypes retrieved from GenBank using the Lasergene software package by the ClustalW method. Phylogenetic and molecular evolutionary analyses were conducted using Mega 4. Genetic distances were estimated using Kimura's 2-parameter method, and standard errors (SE) of the distances were calculated using the bootstrap method (1,000 replicates). Phylogenetic tree was constructed with the Neighbor-Joining method, and its statistical significance was tested using the bootstrap method (1,000 replicates) (Fig. 1).
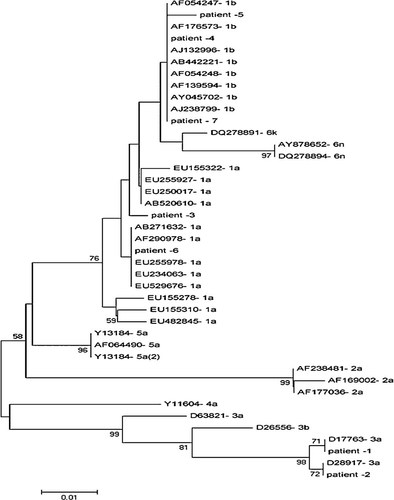
A Neighbor-Joining tree constructed with HCV 5′ non-coding region nucleotide sequences of the clones obtained from the seven patients with occult HCV infection and those corresponding to various HCV genotypes. Bootstrap values ≥70 obtained after 1,000 replicates of the data sheet, are shown in the nodes of the tree.
Immunohistology
Immunohistochemical detection of the HCV core antigen was performed on formalin fixed paraffin-embedded liver biopsy specimens using an anti-HCV core monoclonal antibody (HCV Monotope; ViroStat) at the optimal dilution which was found to be 1:30 [Castillo et al., 2004]. Paraffin was removed by xylene, tissue was rehydrated by immersion in decreasing grades of ethanol and endogenous peroxidase activity was blocked by immersing the tissues in methanol containing 0.3% hydrogen peroxide for 15 min. After antigenic-retrieval (20 min at 95°C in a buffer of citrate, pH = 6), the tissues were incubated overnight (18 hr) at 4°C with primary antibody followed by Dako Envision (1 hr, room temperature) and with addition of aminoethyl carbazole (AEC reagent, Invitrogen, Paisley, UK) as the chromogen. The sections were lightly counterstained with hematoxylin (Dako, Carpinteria, CA) and examined by light microscopy.
This assay was performed on liver biopsy specimens of six patients with occult HCV infection (after diagnosis of them) and two liver biopsy specimens from patients with chronic HCV infection as positive controls.
Statistical Analysis
Data analyses were performed by SPSS ver 16 (SPSS, Chicago, IL). Descriptive analyses as well as Student's t-test were used. A P < 0.05 was considered statistically significant.
RESULTS
A total of 69 patients who were repeatedly found anti-HCV- and serum HCV-RNA-negative and who had abnormal liver function tests of unknown etiology for a mean ± SD 29.7 ± 25.2 months were enrolled in this study. The mean ± SD age of patients was 43.8 ± 14.7 (range: 14–71) years. Out of 69 patients, 48 (71%) were male and 21 (29%) were female. Demographic, laboratory, and epidemiological parameters of patients suffering from chronic liver disease of unknown etiology are summarized in Table I. Table II depicts comparison between patients with and without occult HCV infection.
| Parameters | Value |
|---|---|
| No. of patients | 69 |
| Gender Male: Female | 48:21 |
| Age (year) | 43.8 ± 14.7 (14–71) |
| Duration of diseasea (months) | 69.2 ± 63.1 (13–273) |
| Body mass index (kg/m2) | 24.3 ± 4.5 (16.9–39.1) |
| Laboratory Parameters: | |
| White blood cell | 4,340 ± 1,792 (1,200–9,600) |
| Hemoglobin (g/dL) | 12.8 ± 1.9 (8.3–16.6) |
| Alanine aminotransferase (IU/L) | 71.1 ± 65.1 (17–378) |
| Aspartate transaminase (IU/L) | 78.6 ± 77.6 (27–400) |
| Total bilirubin (mg/dL) | 2.4 ± 2.3 (0.6–17.0) |
| Direct bilirubin (mg/dL) | 0.7 ± 0.9 (0.1–6.9) |
| Serum cholesterol (mg/dL) | 165 ± 36.6 (68–270) |
| Serum triglyceride (mg/dL) | 123 ± 51.4 (30–346) |
| Serum iron (mg/dL) | 74.2 ± 42.0 (8–202) |
| Serum ferritin (ng/dL) | 88.7 ± 141.3 (5–750) |
| Platelet | 95,623 ± 68,661 (12,000–295,000) |
| Prothrombin time (sec) | 16.2 ± 3.3 (12–27) |
| Epidemiological Parameters: | |
| History of surgery | 47 |
| History of blood transfusion | 28 |
| History of endoscopy | 60 |
| History of jaundice | 9 |
- Continuous variables are presented as mean ± SD (range).
- a The estimated duration of abnormal liver function testes since the first alteration was detected.
| Parameters | Positive | Negative | P-value |
|---|---|---|---|
| No. of patients | 7 | 62 | 0.16 |
| Gender Male: Female | 6:1 | 42:20 | |
| Age (year) | 40 ± 13.5 | 44.2 ± 14.9 | 0.23 |
| Duration of diseasea (months) | 71.9 ± 70.4 | 68.5 ± 64.9 | 0.73 |
| Body mass index (kg/m2) | 24.9 ± 4.6 | 24.3 ± 4.5 | 0.86 |
| Laboratory Parameters: | |||
| White blood cell | 4,886 ± 2,490 | 4,277 ± 1,712 | 0.38 |
| Hemoglobin (g/dL) | 11.8 ± 2.5 | 13.0 ± 1.8 | 0.13 |
| Alanine aminotransferase (IU/L) | 107 ± 73.2 | 67.0 ± 63.6 | 0.15 |
| Aspartate transaminase (IU/L) | 99.1 ± 66.1 | 76.2 ± 79.0 | 0.87 |
| Total bilirubin (mg/dL) | 2.3 ± 1.1 | 2.4 ± 2.5 | 0.50 |
| Direct bilirubin (mg/dL) | 0.6 ± 0.3 | 0.8 ± 0.9 | 0.46 |
| Serum cholesterol (mg/dL) | 169 ± 40.2 | 164 ± 36.5 | 0.82 |
| Serum triglyceride (mg/dL) | 123 ± 60.6 | 123 ± 50.9 | 0.49 |
| Serum iron (mg/dL) | 69.4 ± 12.8 | 75 ± 43.8 | 0.05 |
| Serum ferritin (ng/dL) | 147 ± 266.0 | 81.9 ± 120.8 | 0.26 |
| Platelet | 100,290 ± 51,457 | 95,097 ± 70,656 | 0.37 |
| Prothrombin time (sec) | 15.4 ± 2.4 | 16.3 ± 3.4 | 0.54 |
| Epidemiological Parameters: | |||
| History of surgery | 5 | 42 | 0.77 |
| History of blood transfusion | 6 | 22 | 0.001 |
| History of endoscopy | 5 | 55 | 0.18 |
| History of jaundice | 1 | 8 | 0.87 |
- Continuous variables are presented as mean ± SD
- a The estimated duration of abnormal liver function testes since the first alteration was detected.
Genomic HCV-RNA was found by RT-nested PCR assay in PBMCs of 7 (10%) of 69 studied patients, indicating that they had occult HCV infection. Patients with positive PBMC results undertook two additional peripheral blood samplings every 3 months. Their plasma and PBMC specimens were tested again by RT-nested PCR assay, which confirmed the previous positive results. Unfortunately, those patients with negative plasma and PBMC results were not followed.
The results of viral RNA detection by RT-nested PCR assay in plasma and PBMC specimens of patients with chronic HCV infection (positive controls) and healthy blood donors (negative controls) were positive and negative, respectively.
The genotyping of the HCV-RNA isolated from PBMC revealed that 3 (43%) patients with occult HCV infection had genotype 1b, 2 (29%) had genotype 1a, and another 2 (29%) had genotype 3a. The nucleotide sequence analysis of the HCV 5′ non-coding region from all these patients with occult HCV infection confirmed that the patients were infected by genotypes 1b (43%), 1a (29%), and 3a (29%).
A significant (P = 0.001) difference was found in the prevalence of positive HCV-RNA in PBMCs of patients and history of blood transfusion.
None of the liver biopsy specimens from either the patients with occult HCV infection or HCV-positive patients who were included as positive controls were found positive for HCV core antigen, although the experiments were repeated under different conditions including different dilutions, incubation times, and antigen retrieval in order to find the optimal condition.
As mentioned previously, only 23 of 69 patients had a liver biopsy specimen and the rest of the patients had been diagnosed by ultrasonography, clinical presentations, and laboratory results. Therefore, there was no exact data available for comparison of the liver histology between patients with and without HCV-RNA in their PBMCs. However, it is important to point out that liver cirrhosis were detected in four of seven patients with occult HCV infection, and in 52 (84%) of 62 patients without occult HCV infection. However, there were no significant differences (P = 0.26) between these groups.
DISCUSSION
Occult HCV infection is referred to the condition of presence of HCV-RNA in the liver or PBMCs of patients who are negative for anti-HCV antibodies and the genomic HCV-RNA strand in their plasma. In the present study, we demonstrated that 7 (10%) of 69 patients suffering from chronic liver disease of unknown etiology had occult HCV infection, as demonstrated by the presence of HCV-RNA in PBMC specimens using RT-nested PCR assay with primers from 5′ non-coding region of the HCV genome. The HCV genotyping of HCV-RNA isolated from PBMCs revealed that 3 (43%) patients with occult HCV infection had genotype 1b, 2 (29%) had genotype 1a, and another 2 (29%) had genotype 3a. This observation is compatible with the prevalence rate of HCV genotypes reported earlier from Iran [Amini et al., 2009].
Although the gold standard method for the diagnosis of occult HCV infection is the detection of HCV genome in the liver biopsy specimen of patients suffering from chronic liver disease of unknown etiology, testing for HCV-RNA in PBMCs is an alternative procedure when the liver biopsy sample is not available [Barril et al., 2008; Thongsawat et al., 2008]. HCV-RNA could be detected in PBMC specimens of a high percentage (70%) of patients with occult HCV infection [Castillo et al., 2004]. Therefore, it has to be stated that a negative result of RT-nested PCR assay for detection of HCV-RNA in PBMCs does not exclude the presence of HCV genome in the hepatocytes. As a result, it may be possible that the prevalence of occult HCV infection in patients were in fact more than that reported in the present study.
There are some reports indicating that all patients with occult HCV infection had HCV genotype 1b in their liver biopsy specimens [Castillo et al., 2004; Pardo et al., 2006] and in PBMCs [Barril et al., 2008]. It should be noted that in these populations, the percentage of HCV genotype 1b is >90% of chronic HCV infections [Quiroga et al., 1998; Rodriguez-Inigo et al., 1999; Laskus et al., 2007]. This high rate of genotype 1b reported in such studies might be due to the high prevalence of this genotype in their populations (Spain). There is a recent report indicating that different HCV genotypes (1a, 1b, 2a) were detected from PBMC samples of patients with occult HCV infection, however, another study from Italy showed that the most prevalent HCV genotype is 1b [De Marco et al., 2009]. HCV genotypes 1a, 6f, and 1b were also isolated from PBMC samples taken from patients with occult HCV infection during an outbreak in a hemodialysis unit in Thailand [Thongsawat et al., 2008], while the most prevalent HCV genotype in this country is 3a (50%–60%) with genotypes 1a, 1b, and 6 comprising the rest (10%–20% each) [Sy and Jamal, 2006].
We know that in our population, the most prevalent HCV genotypes are 1a (39.7%), 3a (27.5%), and 1b (12.1%) [Keyvani et al., 2007]. These percentages are not completely similar to the frequencies of HCV genotype in PBMC specimens taken from the patients with occult HCV infection in the present study.
On the other hand, there are some reports which demonstrated that the frequency of HCV genotypes existing in PBMCs was different from those found in plasma or liver biopsy specimen [Roque-Afonso et al., 2005; Bokharaei Salim et al., 2010]. Therefore, it seems that prospective studies focusing on identification of HCV genotypes involved in occult HCV infection in the general population will be needed.
HCV is among the infectious agents that can be transmitted through blood transfusion; although the risk of transmission of infectious agents through transfusion can be estimated using the prevalence of the disease conditions in donors' population, screening level for infection disease, and sensitivity and specificity of the tests used [Schmunis et al., 1998]. In the present study, a significant difference was observed in the history of blood transfusion between patients with and without occult HCV infection. Therefore, patients with occult HCV infection may acquire the infection from blood transfusion since despite advances in screening tools, the risk of transmission through blood transfusion still exists [De Marco et al., 2009]. On the other hand, the risk of HCV transmission from patients with occult HCV infection via blood transfusion should be considered, because despite the absence of a detectable genomic HCV-RNA in serum of these subjects, they could be potentially infectious through their PBMCs.
Our immunohistochemical analysis did not detect HCV core antigen in none of the liver biopsy specimens from patients either with occult HCV infection or HCV positive patients who were used as positive control group. These findings are in line with previous studies which showed that the levels of expression HCV antigens in liver may be undetectable by standard immunohistichemistry method [Yap et al., 1994; Sansonno et al., 1995; Brody et al., 1998; Castillo et al., 2004].
In conclusion, we have demonstrated in the present study that in our population, more than 10% of patients suffering from chronic liver disease of unknown etiology have occult HCV infection. Therefore, the possibility of this infection should be considered in those patients who suffer from chronic liver disease of unknown etiology.




