Monoclonal antibodies to the haemagglutinin HA1 subunit of the pandemic influenza A/H1N1 2009 virus and potential application to serodiagnosis
Abstract
In order to provide specific serological reagents for pandemic influenza A/H1N1 2009 virus, monoclonal antibodies (Mabs) to recombinant haemagglutinin component HA1 (rHA1) were generated after fusing spleen cells from a mouse immunized with rHA1 protein derived from influenza strain A/California/06/09 H1N1 with a mouse myeloma cell line. Five hybridoma clones secreting Mabs specific for the rHA1 protein derived from pandemic influenza A/H1N1 2009 and not for rHA1 from seasonal H1N1 influenza strains A/Brisbane/59/07 and A/Solomon Islands/03/06 were identified by EIA. Mabs 7H4, 9A4, and 9E12 were reactive in Western blots with full length rHA and/or rHA1 subunit derived from A/California/06/09 strain. Only Mab 1F5 inhibited haemagglutination of turkey red blood cells with recombinant NIBRG-121 virus derived from A/California/07/09, but did not react in Western blots. Immunostaining of MDCK cells infected with NIBRG-121 was localized to the membrane/cytoplasm for four of the reactive Mabs. The differing reactivity of the Mabs in Western blots, immunostaining, EIA, and haemagglutination inhibition assay suggest that at least four of the five Mabs recognize different epitopes on HA1 of the pandemic influenza A/H1N1 2009 virus. Ferret antisera to pandemic influenza A/H1N1 2009 (A/England/195/09 and A/California/07/09 strains) and sera from human subjects vaccinated with Influenza A (H1N1) 2009 Monovalent Vaccine (CELTURA®, Novartis Vaccines, Germany), inhibit binding of 1F5-HRP to biotinylated rHA1 derived from A/California/06/09 in a competitive EIA, suggesting that the epitope recognized by this Mab also evokes an antibody response in infected ferrets and vaccinated humans. J. Med. Virol. 83:559–567, 2011. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Influenza A viruses are negative sense, single-stranded RNA viruses that belong to the genus Influenzavirus A in the family Orthomyxoviridae. The haemagglutinin (HA) of influenza resides on the surface of the virus and initiates the viral infectious cycle by attaching to sialic acid residues in host cells and inducing fusion (Skehel and Wiley, 2000). Depending on the virus strain, host cell type, and growth conditions used, the HA can exist in its uncleaved form (HA0 MW ∼ 76 kDa) or in its cleaved form, consisting of two di-sulfide linked chains HA1 and HA2 (Klenk et al., 1975; Lazarowitz and Choppin, 1975). HA is the major viral antigen against which neutralizing antibodies are produced, and influenza epidemics are associated with changes in its antigenic structure (Lamb and Krug, 1996). Influenza pandemics occur when an influenza virus with a HA, against which there is little or no existing immunity, emerges in human populations and transmits efficiently from human to human (Garten et al., 2009). Between March and April 2009, a previously unknown variant of influenza A/H1N1 caused sporadic infections and epidemics in North America and Mexico, resulting in a global pandemic (Hancock et al., 2009; MMWR, 2009a,b; Miller et al., 2010; Ikonen et al., 2010). Antigenic and genetic characterization of the 2009 pandemic influenza A/H1N1 virus revealed it to be very different from seasonal H1N1 viruses (Garten et al., 2009). Structural analysis revealed up to 27% difference between the amino acid sequence of HA1 of the pandemic virus influenza A/H1N1 2009 and the 2007 seasonal influenza A H1N1 virus, with the majority of the changes in the globular, antigenic sites of the HA1 protein (Ikonen et al., 2010). Consequently, serological reagents specific for seasonal influenza H1N1 cannot be used for detection of the pandemic influenza A/H1N1 2009 virus.
The aim of the present study was to generate monoclonal antibodies (Mabs) specific for the HA1 of pandemic influenza A/H1N1 2009 strain and to characterize these antibodies in terms of specificity, potential utility in research, and serological investigations of the influenza A/H1N1 2009 pandemic.
MATERIALS AND METHODS
Viral Proteins
The following purified recombinant proteins, all of which were 6x His tagged and expressed in 293 cells, were obtained from e-Enzyme LLC (Montgomery Village, MD, USA): haemagglutinin HA1 (A California/06/09 H1N1; amino acids 18–344), haemagglutinin HA (A/California/06/09 H1N1; amino acids 18–530), haemagglutinin HA1 (A/Brisbane/59/07 H1N1; amino acids 18–343), haemagglutinin HA (A/Brisbane/59/07 H1N1; amino acids 18–530), haemagglutinin HA1 (A/Solomon Islands/3/06 H1N1; amino acids 18–344), haemagglutinin HA (A/Solomon Islands/3/06 H1N1; amino acids 18–530), haemagglutinin HA1 (A/New Caledonia/20/99 H1N1; amino acids 18–345), haemagglutinin HA (A/New Caledonia/20/99 H1N1; amino acids 18–530), HA (B/Malaysia/2506/04; amino acids 12–547), haemagglutinin HA1 (A/Vietnam/1203/04 H5N1; amino acids 1–345) and haemagglutinin HA (A/Vietnam/1203/04 H5N1; amino acids 18–530).
Biotinylation of Proteins
Approximately 50 µG of the viral proteins and 6 mG of bovine serum albumin (BSA) were biotinylated using the EZ-Link™NHS-Biotin from Pierce (Rockford, IL, USA) essentially as described in the supplied instructions.
Streptavidin Coated Microtiter Plates
12 × 8 U-well strip microplates, (Greiner Bio-one, Stonehouse, UK) were first coated with 100 µL/well of a 50 ng/mL solution of BSA-biotin in 0.05 M carbonate buffer, pH 9.5 at 2–8°C overnight. The plates were washed with PBS containing Tween-20 (0.05% v/v, PBST) and then blocked with 200 µL/well of a blocking buffer (Microimmune Ltd, Hounslow, UK) for 2 hr at 37°C. To the BSA-biotin plates was added 500 ng/mL streptavidin (Prozyme, Inc, Hayward, CA, USA) in the blocking buffer for 1 hr at 37°C and the wells aspirated and stored at 2–8°C until used.
Monoclonal Antibody Production
A BALB/c mouse was immunized subcutaneously at fortnightly intervals on four separate occasions with a mixture containing 40 µg of rHA1 (A/California/06/09) protein in 90 µL of PBS containing 50 µg of N-acetylmuramyl-L-alanyl-D-isoglutamine hydrate (MDP, Sigma–Aldrich Co. Ltd, Poole, UK) and an equal volume of Freund's Incomplete Adjuvant (Sigma–Aldrich Co. Ltd). A test bleed was taken one week after the last immunization. Three days prior to splenectomy, the mouse was again immunized subcutaneously as before, except that 25 µg of rHA1 was used.
The splenocytes from the immunized mouse were fused with the NS1 mouse myeloma cell line by standard procedures described previously (Kohler and Milstein, 1976; Tedder et al., 1982) and the fused cells distributed into 10 × 96-well tissue culture plates containing selective hypoxanthine, aminopterin, and thymidine medium. The serum obtained from the test bleed was used to develop the enzyme immunoassays (EIAs) used for sceening hybridoma supernatants described below. Hybridomas found to be positive in the EIA were expanded and cloned by limiting dilution.
EIA for Screening of Hybridoma Supernatants
Separate microtiter plates (Greiner Bio-one) were coated with rHA1 subunit (A/California/06/09) and whole rHA (A/California/06/09) in carbonate coating buffer as described for streptavidin coated plates. In addition, biotin-rHA1 plates were prepared by adding 100 µL/well of biotinylated rHA1 (200 ng/mL) in 3% (w/v) BSA in PBST to the streptavidin coated plates and incubating at 2–8°C overnight. After washing with PBST, these wells were also used to screen hybridoma supernatants in the EIA procedure described below.
To 100 µL of hybridoma supernatants was added 250 µL of 1% (w/v) milk in PBST containing 0.05% (w/v) Bronidox-L (Henkel, Germany, PBSTBx). One hundred microliters of the diluted hybridoma supernatant was added to wells of rHA1 (A/California/06/09), rHA (A/California/06/09) and biotin-rHA1 (A/California/06/09) coated plates and incubated for 1 hr at 37°C. The plates were washed four times with PBST and 100 µL/well of rabbit anti-mouse IgG-horseradish peroxidase (HRP) conjugate (Dako UK Ltd, Ely, UK), diluted 1/4 000 in 1% (w/v) milk in PBSTBx, was added and incubated for 30 min at 37°C. The plates were washed and incubated with 100 µL/well of TMB substrate (Microimmune, Hounslow, UK) until color developed. The color development was terminated by the addition of 100 µL/well of 0.5 M hydrochloric acid and the optical density readings at 450/620 nm in each well recorded.
Isotyping of Monoclonal Antibodies
Goat anti-mouse IgG1, IgG2a, IgG2b, IgG2c, IgG3, and IgM were obtained from Jackson Immunoresearch Laboratories (Stratech Scientific Ltd, Newmarket, UK). Tissue-culture supernatants (100 µL) from cloned hybridomas were added to immobilized biotin-rHA1 (A/California/06/09) wells as described above and incubated for 1 hr at 37°C. After washing the wells as before in PBST, 100 µL of a 1/2 000 dilution of each of the goat-anti-mouse isotyping reagents in 1% (w/v) milk in PBSTBx was added and incubated for 30 min at 37°C. The wells were washed as before and incubated with 100 µL of a 1/20 000 rabbit anti-goat IgG-HRP conjugate (Sigma–Aldrich Co. Ltd.) in 1% (w/v) milk in PBSTBx and incubated for 30 min at 37°C. The bound HRP was detected with TMB substrate as described above.
EIA for the Quantitation of IgG in Culture Supernatants
Microtiter plates were coated with 100 µL/well of rabbit anti-mouse IgG (5 µg/mL, Sigma, Dorset, UK) in 0.05 M carbonate buffer, pH9.5 overnight at 2–8°C. The wells were washed and 100 µL of dilutions of IgG (derived from clone 1F5) in 1% (w/v) milk in PBSTBx was added to the wells in triplicate for construction of a standard curve. Simultaneously 100 µL of Mab culture supernatants, diluted in the same buffer, were added in triplicate to separate wells and incubated at 37°C for 1 hr. The plates were washed as before and 100 µL of a 1/4 000 dilution of rabbit anti-mouse IgG-HRP conjugate was added. After washing the wells four times with PBST, the bound HRP was detected with TMB substrate.
SDS PAGE and Western Blotting
Recombinant viral proteins were separated electrophoretically under reducing conditions on a NuPAGE 10% Bis-Tris gel (Invitrogen Ltd, Paisley, UK) using MOPS running buffer and the electrophoresed proteins transferred onto nitrocellulose membranes essentially as described by Towbin et al., (1979) and detailed in the product literature from Invitrogen Ltd. The membranes were then incubated with a 1/20 dilution of supernatants derived from each of the cloned Mab producing cell lines in 1% (w/v) milk in PBST or with a 1/100 dilution of serum from the mouse test bleed in the same buffer solution for 1 hr at room temperature (RT). The membranes were then washed and incubated with a 1/20 000 dilution of a rabbit anti-mouse IgG-alkaline phosphatase conjugate (Stratech Scientific Ltd). After washing, the bound Mabs were detected with 5-bromo-4-chloro-3-indolyl phosphate and nitro-blue tetrazolium (NBT/BCIP) as substrate (Leary et al., 1983).
Haemagglutination Inhibition (HAI)
The ability of the Mabs to inhibit agglutination of 0.5% turkey red blood cells (RBC) by two recombinant influenza A virus strains NIBRG-121 and NIBRG-122, derived from A/California/07/09 and A/England/195/09, respectively, and supplied by the National Institute for Biological Standards and Controls (NIBSC), UK, was tested essentially as described earlier (Ellis and Zambon, 1997). Briefly, supernatants were diluted two-fold in PBS, starting at a 1:8 dilution, and 4 haemagglutination units of virus were added, followed by incubation for 1 hr at RT, after which the turkey RBC were added. Results were interpreted after 30 min incubation at RT.
Cell-ELISA and Immunostaining of Virus Infected MDCK Cells
Madine-Darby canine kidney (MDCK) cells were infected with recombinant virus NIBRG-121 at a multiplicity of infection of 0.05 particles per cell and dispensed into wells of 96-well tissue culture plates using standard procedures (Rowe et al., 1999). One hundred microliters of diluted Mab supernatants in 1% (w/v) milk PBSTBx were added to the wells and incubated for 1 hr at RT. The wells were washed four times by gentle aspiration of the antibody solution and washing with PBST. The wells were incubated with 100 µL rabbit anti-mouse IgG-alkaline phosphatase conjugate diluted 1/20 000 in 1% (w/v) milk PBSTBx or with 100 µL of rabbit anti-mouse IgG-HRP conjugate diluted 1/4 000 in the same buffer. The wells were washed four times with PBST as before and further washed twice with water before immunostaining. The HRP conjugates were revealed by the addition of 100 µL of a soluble TMB substrate for the cell-ELISA and stopping the reaction after color development with 100 µL 0.5 M HCl and reading the optical density at 450/620 nm. For localization of Mab staining, the HRP conjugates were visualized after addition of 100 µL of a precipitating substrate, TrueBlue™, (KPL, Gaithersburg, MD, USA) and the alkaline phosphatase conjugates were revealed with the addition of 100 µL/well of NBT/BCIP substrate, as described for Western blotting above.
Competitive EIA
Supernatant from Mab 1F5-hybridoma cell-culture was harvested and IgG purified using protein A column and labeled with horseradish peroxidase using standard procedures (Ishikawa et al., 1983; Nilsson et al., 1981). The purified conjugate contained 1.44 moles of enzyme per mole of IgG and the antibody concentration was approximately 2 mg/mL. To streptavidin coated plates (see above) 100 µL/well of a 100 ng/mL biotin-rHA1 from A/California/06/09 in 1% (w/v) milk PBSTBx was added and incubated at 37°C for 1 hr. The plates were then washed four times in PBST.
An equal volume of HRP conjugated 1F5 Mab (1F5-HRP) diluted 1/1 000 was mixed with an equal volume of competing antibody. The competing antibodies were undiluted Mab supernatants, or 1/5 dilutions of ferret antisera to influenza A virus strains (see Fig. 3a for strains) and serum samples from humans pre and post immunization with Celtura® vaccine (containing the inactivated surface antigen preparation of the reassortant NYMC X-179A virus) in 1% (w/v) milk PBSTBx. One hundred microliters per well of this was then added in duplicate to streptavidin-biotin-rHA1 wells for 1 hr at 37°C. After washing, the plates were incubated with TMB substrate (Microimmune Ltd.) at RT for 20 min. Known concentration of unconjugated antibody from clone 1F5 was used as the competing antibody to generate a standard curve. Four or eight replicate control wells, in the absence of competing culture supernatant or sera from ferrets and human vacines, respectively, were used to calculate the mean OD450/620nm of binding by 1F5-HRP. The percent inhibition was calculated using the equation: (mean OD450/620nm 1F5-HRP—mean OD450/620nm competing supernatant or serum sample)/mean OD450/620nm 1F5-HRP × 100. Where competing serum samples gave OD450/620nm values greater than the control in the absence of serum, the result was designated a value of 0% inhibition.
Experimental Ethics
All experimental work involving animals were performed in compliance with the Animals (Scientific Procedures) Act1986 under licence and in accordance with ethical standards of the Declaration of Helsinki.
RESULTS
EIA Screening of Hybridoma Colonies
Fourteen days post fusion virtually all of the 960 wells from the fusion plates had colonies of hybridoma growth and consequently, supernatant from all the wells were screened by EIA. Three strategies were used for the screening procedure. Reactivity of the hybridoma supernatant to rHA or rHA1 derived from H1N1 (A/California/06/09) coated directly on to microtiter wells or to biotinylated rHA1 derived from H1N1 (A/California/06/09), immobilized using streptavidin coated plate. The serum derived from the test bleed of the immunized mouse gave strong EIA reactivity in all three assays. The five monoclonals taken forward all reacted with biotin-rHA1 coated wells. Only one of these Mabs reacted with rHA1 coated directly onto microtiter wells. The EIA positive hybridoma cells were cloned by limiting dilution.
EIA for the Estimation of IgG Concentration Culture Supernatants
From a standard curve constructed using purified mouse IgG1, the concentration of IgG in the culture supernatants used in the characterization of Mabs was estimated. The concentrations of antibody in the supernatant for Mabs 1F5, 9A4 and 7H4 were between 15–18 µg/mL, and for supernatant 9E12, was much higher (26.0 µg/mL). The antibody concentration in the supernatant of Mab 1G10 was only 1.0 µg/mL (see Table I).
| Monoclonal antibody | Reactivity in EIA (OD 450/620nm) | HAI titer | Mab reactivity with MDCK cells infected with NIBRG-121 virus in: | Reactivity in Western blot | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone designation | Isotype | Conc. µg/mL | Biotin-rHA1 subunit A/California/06/09 | Biotin-whole rHA A/California/06/09 | Biotin rHA1 subunit A/Solomon Is./03/06 | Biotin rHA1 subunit A/Brisbane/59/07 | NIBRG 121 | NIBRG 122 | Cell-ELISA HRP:TMB OD 450/620nm | NBT-BCIP | True-Blue | Whole rHA | Subunit rHA1 |
| 1F5 | IgG1 | 16.1 | 0.555 | 0.357 | 0.017 | 0.018 | 128 | 16 | 1.018 | +++ | +++ | − | − |
| 1G10 | IgG1 | 1.0 | 0.056 | 0.050 | 0.011 | 0.013 | <8 | <8 | 0.177 | +/− | + | − | − |
| 7H4 | IgG1 | 17.7 | 0.248 | 0.098 | 0.014 | 0.018 | <8 | <8 | 0.095 | − | − | +++ | +++ |
| 9A4 | IgG1 | 15.2 | 1.138 | 0.569 | 0.013 | 0.016 | <8 | <8 | 0.718 | +++ | +++ | +++ | +/− |
| 9E12 | IgG1 | 26.0 | 0.597 | 0.108 | 0.009 | 0.011 | <8 | <8 | 0.170 | +/− | + | − | + |
- NIBRG-121 and NIBRG-122, are recombinant viruses derived from influenza A/California/07/09 and A/England/195/09 strains, respectively.
- Intensity of staining of MDCK cells infected with NIBRG-121 virus using insoluble substrates, NBT-BCIP (for alkaline phosphatase conjugate) and True-Blue™ (for HRP conjugate), by each Mab supernatant was scored as strong (+++), moderate (+), weak, (+/−) or not detected (−). Intensity of staining of whole rHA or rHA1 subunit, derived from influenza A/California/06/09 H1N1 virus, in Western blot was scored as for immunostaining of MDCK cells infected with NIBRG-121.
Characterization of the Monoclonal Antibodies
Since all five clones reacted with biotinylated rHA1 subunit derived from A/California/06/09, we assessed their reactivity with biotinylated rHA A/California/06/09 and with biotinylated rHA1 subunit derived from A/Brisbane/07/09 H1N1 and A/Solomon Islands/03/06 H1N1 in EIA using streptavidin to capture the biotinylated proteins. All five clones were specific for rHA and rHA1 derived from A/California/06/09 and did not react in EIA with the biotinylated recombinant proteins from A/Brisbane/07/09 or with A/Solomon Islands/03/06. Serum from the test bleed reacted with all four biotinylated proteins. Isotyping of the Mab supernatants showed that all five clones secreted IgG1 antibodies (Table I).
In Western blots, serum from the test bleed of the immunized mouse, used to generate the Mabs, stained all the recombinant haemagglutinin proteins derived from H1N1 influenza strains, but not the recombinant haemagglutinin form H5N1 and influenza B (Fig. 1a). Mabs from clones 1F5 and 1G10 did not react with any of the proteins in Western blots, although the lack of staining observed for 1G10 may be explained by its low antibody concentration in culture supernatants. Mab 7H4 gave intense immunostaining with rHA and rHA1 from A/California/06/09, but did not stain any other recombinant protein (Fig. 1b). Mab 9A4 gave intense reaction with rHA from A/California/06/09, but only weak staining with rHA1 from A/California/06/09 (Fig. 1c). Mab 9E12 showed weak reaction with rHA derived from A/California/06/09 only (Fig. 1d).
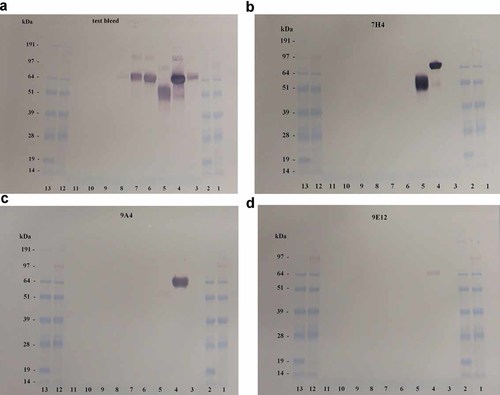
Western blot analysis of test bleed and monoclonal anti-HA1 antibodies. Recombinant HA proteins derived from influenza strains were analyzed using SDS-PAGE under reducing conditions, transferred to nitrocellulose and immunostained with (A) serum from test bleed and Mabs (B) 7H4 (C) 9A4 and (D) 9E12. Lanes 1 and 12: SeeBlue® Plus2 molecular weight standard, Lane 2 and 13: SeeBlue® molecular weight standard, Lane 3: HA (A/New Caledonia/20/99 H1N1), Lane 4: HA (A/California/06/09 H1N1), Lane 5: HA1 (A/California/06/09 H1N1), Lane 6: HA (A/Solomon Islands/3/06 H1N1), Lane 7: HA (A/Brisbane/59/07 H1N1), Lane 8: HA (A/Vietnam/1203/04 H5N1), Lane 9: HA1 (A/Vietnam/1203/04 H5N1), Lane 10: HA (B/Malaysia/2506/04), Lane 11: Blank.
In a cell-ELISA, all five Mabs reacted with the recombinant virus NIBRG-121, with differing intensities (See Table I). Mab 1F5 and 9A4 gave the highest OD signals using TMB substrate. Diffuse cytoplasmic/membrane staining of infected cell was observed with Mabs 1F5 and 9A4 (Fig. 2a,b) using both the insoluble substrates, NBT/BCIP substrate (with alkaline phosphatase conjugate) and TrueBlue™ (with HRP conjugate). This was different from the weak cytoplasmic/membrane staining pattern observed with 9E12 (Fig. 2c). The weak staining of infected cells by 1G10 may be the result of the low concentration of antibody in the culture supernatant used (Fig. 2d).
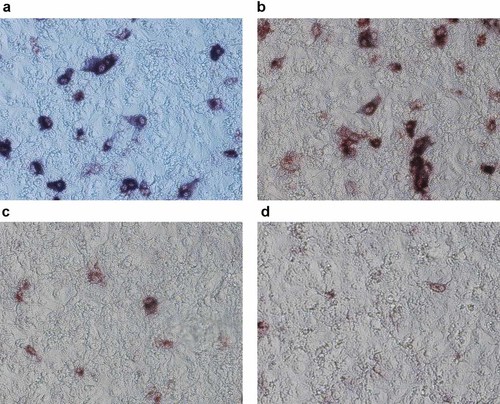
MDCK cells infected with recombinant NIBRG-121 virus immunostained using monoclonal antibody and anti-mouse alkaline phosphatase conjugate. Staining of infected cells with (A) Mab 1F5 (B) Mab 9A4 (C) Mab 9E12 and (D) Mab 1G10 is shown.
Only Mab 1F5 inhibited haemagglutination of turkey RBC by recombinant virus NIBRG-121 (derived from A/California/07/09) and recombinant NIBRG-122 (derived from A/England/195/09). The HAI titer of 1F5 with NIBRG-121 was eight-fold higher than with NIBRG-122 (Table I).
In competitive EIA, only serum from the test bleed and the unconjugated tissue culture supernatant from clone 1F5 competed effectively with 1F5-HRP labelled antibody. A standard curve for the competitive EIA using purified, unconjugated 1F5 antibody revealed that the detection limit was 75 ng/mL. Partial inhibition of 1F5-HRP binding in EIA was observed with tissue culture supernatants Mab 9A4 (38% inhibition) and 9E12 (12%). No inhibition was observed with supernatants from Mab 7H4 and 1G10.
The inhibition of 1F5-HRP with ferret sera in the competitive EIA is shown in Figure 3a and the corresponding HAI results with NIBRG-121 are shown over the bars in the graph where available. Two ferrets immunized with the same, recent European swine-lineage influenza strain (A/Aragon/3218/08; provided by the Director of the National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain) showed between 10% and 16% inhibition of 1F5-HRP in the competitive EIA, whereas ferrets immunized with the 2009 pandemic influenza strain A/California/07/09 H1N1 and influenza A/England/195/09 H1N1 showed approximately 55% and 30% inhibition of conjugate binding, respectively. Of the ferret antisera to recent seasonal influenza A/H1N1, only the A/Brisbane/59/07 showed any significant inhibition of 1F5-HRP binding (7.7%) in the competitive EIA. All other ferret antisera failed to inhibit 1F5-HRP binding. The ferret antisera to the two pandemic influenza A/H1N1 2009 virus strains had high HAI titers with NIBRG-121. Significant but lower HAI titers were obtained for ferret antisera to the recent swine-lineage virus A/Aragon/3218/08 and to antisera to the strain A/swine/England/117316/86.
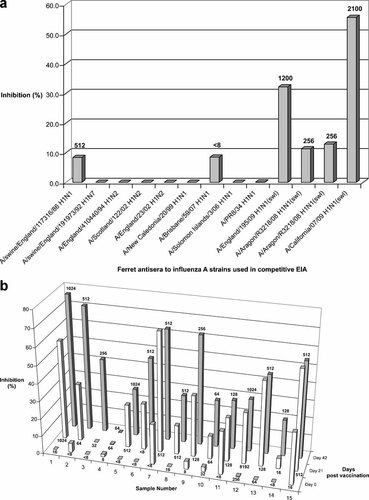
a: Inhibition by ferret antisera of Mab 1F5-HRP binding in a competitive EIA. The antisera to the influenza A strains are identified on the abscissa and the per inhibition of 1F5-HRP binding for each of these is shown on the ordinate. The numbers shown above the bars in the graph are the corresponding HAI titers obtained for each antiserum with NIBRG-121. b: Inhibition by pre and post vaccination sera of Mab 1F5-HRP binding in a competitive EIA. The HAI titers for each subject pre and post vaccination with Celtura® vaccine are shown above or at the base of the individual bars in the graph.
The inhibition of 1F5-HRP binding obtained with sera from 15 subjects pre and post vaccination with Celtura® adjuvanted pandemic H1N1 vaccine is shown in Figure 3b and the HAI titers obtained for each of the specimens is given on the bar chart. Twelve of the 15 sera taken on the day the vaccine was administered (day 0) did not inhibit 1F5-HRP binding. This included five sera with HAI titers of 16 or greater on day 0. Serum from one subject in particular (subject 12) had an HAI titer of 256 on day 0, but did not show any inhibition of 1F5-HRP in the competitive EIA. Of the remaining three sera collected on day 0, two had HAI titers of <8 and one of 16 (subjects 7, 11, and 15) and these inhibited 1F5-HRP (inhibition of 23%, 24%, and 29%, respectively). Most subjects had seroconverted by HAI and also produced antibodies that inhibited 1F5-HRP binding in the competitive EIA by Day 21, with two exceptions, subjects 3 and 4. Subject 3 showed seroconversion in the competitive EIA on day 42, whereas subject 4 remained seronegative by competitive EIA at this time and maintained an HAI titer of 64, as observed on day 21. In general, sera from subjects giving high percent inhibition of 1F5-HRP in the competitive EIA also had high HAI titers, although there were some exceptions (see subjects 12 and 6 taken on day 21). The correlation between HAI titers and percent inhibition in the competitive EIA was poor (R2 < 0.3).
DISCUSSION
An operational consequence of the emergence of the pandemic influenza A/H1N1 2009 was that the pathogen and specimens from suspected cases needed to be handled at a higher biological containment level than for seasonal influenza viruses. This restricted the propagation of virus in cell culture to laboratories with high containment facilities and further restricted availability of the virus or viral proteins for developing serological reagents. However, within three months of the first cases of the pandemic influenza A/H1N1 2009 virus being reported in Mexico and the USA, rHA proteins derived from the pandemic strain in purified form became available commercially. This prompted us to generate Mabs to rHA in order to provide additional serological research tools for studying the pandemic influenza A/H1N1 2009 virus strains.
Recombinant HA1 derived from A/California/06/09 H1N1 proved to be immunogenic, since serum taken from the test bleed of the immunized mouse showed reactivity with rHA1 subunit and whole rHA (derived from the A/California/06/09 strain) coated wells and with biotinylated rHA1 from the same strain suspended from streptavidin coated wells in EIA. The majority of hybridomas screened did not react with the recombinant proteins coated directly onto polystyrene wells. This may have occurred as a result of failure in the fusion process to fuse splenocytes with specificity for the proteins. However, success with using biotinylated rHA1 suspended from streptavidin coated plates for identifying reactive hybridomas suggested that conformational presentation of the rHA protein was important. It may be that the rHA1 and rHA undergo changes in conformation on binding to polystyrene wells, as was shown for HA binding to polyvinyl by Yewdell (2010). Yewdell (2010) demonstrated recently that some Mabs to discontinuous epitopes on influenza A HA can induce refolding of denatured proteins bound to polyvinyl plates over a two day incubation period. Thus it is possible that with extended incubation of the hybridoma supernatant during the screening process, more hybridoma clones reactive to conformational epitopes in rHA1 may have been identified.
Four of the five Mabs to the pandemic H1N1 influenza A/California/06/09 generated in this study appeared to recognize different epitopes as demonstrated by the differing pattern of reactivity observed in Western blots, staining of virus infected cells, HAI, and in competitive EIA. The concentration of IgG in the culture supernatant of Mab 1G10 was 10 times lower than that found for other Mabs and it was therefore difficult to establish if this Mab recognized a unique epitope. All Mabs were specific for the pandemic influenza A/H1N1 2009 strain and did not cross-react with pre-2009 seasonal H1N1 strains. Unlike the Mabs used in this work, which proved to be specific for A/California/06/09 in EIA and Western blots, the polyclonal serum from the test bleed mouse showed cross-reactions with the HA1 of influenza A/Brisbane/59/07 and influenza A/Solomon Islands/03/06 and A/New Caledonia/2/99. This is not surprising since there is ∼70% amino acid sequence homology between the HA1 of A/California/06/09 and seasonal influenza A/H1N1 (Ikonen et al., 2010).
There are only three amino acid changes in the haemagglutinin between A/California/07/09 and A/England/195/09 (L49I, P91S, and I323V) and this was sufficient to result in an eight-fold difference in HAI titers of 1F5 seen with the two recombinant viruses. Furthermore, in competitive EIA there was greater inhibition of 1F5-HRP binding by ferret antiserum to A/California/06/09 compared to A/England/195/09. Although the observed differences with the animal sera may be due to differing responses to infection in individual ferrets, the combined HAI and competitive EIA data suggest that the amino acid changes in the two haemagglutinins result in a significant change in the epitope recognized by the 1F5 antibody. If there are significant changes in the IF5-epitope in other 2009/10 virus isolates, then Mab IF5 could prove useful for tracking antigenic drift in the pandemic influenza A/H1N1 2009 virus strains.
In the competitive EIA, inhibition of 1F5-HRP binding to the biotinylated rHA1 of A/California/06/09 would only be expected if competing antibodies either compete directly for the epitope recognized by Mab 1F5 or if the competing antibodies bind to other epitopes on the rHA1 that are close enough to interfere sterically with 1F5-HRP binding. Since Mab 1F5 inhibits haemagglutination of turkey RBC by NIBRG-121, it would follow that any antibodies that inhibit 1F5-HRP in the competitive EIA as a result of steric interference will inhibit haemagglutination. Although this proved to be the case for most of the ferret antisera and post vaccination sera tested (Fig. 3a,b), the correlation between the HAI titers and % inhibition observed in the competitive EIA was poor. This is not surprising, since HAI measures a polyclonal antibody response directed toward a number of antigenic epitopes on the haemagglutinin receptor binding site of the virus, whereas the competitive EIA only measures antibodies in the competing serum that inhibit binding of a single epitope on haemagglutinin of the virus. Although 1F5 inhibits haemagglutination of NIBRG-121, it does so through binding to a single epitope on the virus, albeit represented multiple times on the virion. Ferret antisera and immune human sera effect haemagglutination by binding to multiple epitopes, represented multiple times on the virion.
An alternative explanation for the poor correlation between HAI and the competitive EIA may be that antibodies which do not inhibit HAI may nevertheless bind to distal sites on the biotinylated rHA1 and cause conformational changes. These conformational changes may have two opposing effects in the competitive EIA. It may improve presentation of the 1F5 epitope, augmenting the signal obtained in the competitive EIA as a result of increased binding of 1F5-HRP, as was observed with some of the sera tested (data not shown). Conversely, an induced conformational change may have an adverse affect on the 1F5 epitope structure, inhibiting binding of 1F5-HRP to the biotinylated rHA1.
Our data on a limited number of subjects investigated in this work indicates that vaccination with Celtura® vaccine induces an immune response to the haemagglutinin and specifically to an epitope shared by 1F5 antibody or to epitopes close to this site. It would be of interest to determine whether individuals infected with the pandemic influenza A/H1N1 2009 virus also mount a significant antibody response to the 1F5-epitope or epitopes close to this site. If further studies show this to be the case then the competitive EIA described here could provide a potentially useful and specific sero-epidemiological tool for studying exposure to infection with the pandemic virus as well as to determine efficacy of vaccination. The competitive EIA has the added benefit that it is simple, safe and not as work-intensive as the haemagglutination inhibition and microneutralization assays.
Acknowledgements
We thank Dr. Joanna Ellis and Dr. Monica Galliano for providing information on amino acid sequence data for the HA1 of 2009 pandemic and seasonal influenza A/H1N1 viruses. We also acknowledge Surita Gangar, Janice Baldevarona and Aram Afsar for technical assistance with haemagglutination inhibition and culture of MDCK cells.




