Human bocavirus infection in children with acute gastroenteritis in Japan and Thailand
Abstract
A total of 329 fecal specimens, which had been known to be negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus, and which were collected from infants and children with acute gastroenteritis in Japan and Thailand during 2005–2008 were screened for human bocavirus (HBoV). HBoV was detected by PCR with a primer pair that amplified the NP1 region of its genome and was genotyped by sequencing of the VP1/VP2 region. Of the 329 samples tested, 6 (1.8%) were positive for HBoV. Of these, five samples were collected from Japan and one sample was from Thailand, and the detection rates of HBoV in each country were 2% and 1.2%, respectively. For the detected HBoV, the capsid VP1/VP2 gene of all HBoV strains was successfully sequenced. Four Japanese HBoV strains studied were clustered into group 1, while the remaining Japanese strain and a unique Thai strain belonged to group 2. No severe acute gastroenteritis associated with HBoV was noted. This study provides better understanding on the epidemiology of HBoV infections in children with acute gastroenteritis in Japan and Thailand. J. Med. Virol. 83:286–290, 2011. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Acute gastroenteritis is one of the most common diseases in infant and children and continues to be a significant cause of morbidity and mortality worldwide [Musher and Musher, 2004]. Rotavirus, calicivirus, astrovirus, and adenovirus have been established as the most important etiologic agents [Glass et al., 2001; Marie-Cardine et al., 2002]. However, the etiologic agents of more than a half of patients with acute gastroenteritis are undiagnosed [Simpson et al., 2003; Olesen et al., 2005; Chanit et al., 2009]. In 2005, human bocavirus (HBoV) was first identified in pooled human respiratory tract specimens from Swedish children with respiratory tract disease and classified into the genus Bocavirus of the subfamily Parvovirinae, family Parvoviridae [Allander et al., 2005]. The HBoV genome is approximately 5.2 kb nucleotides in length and contains three open-reading frames (ORF) encoding two non-structural proteins (NS1 and NP-1) and two capsid proteins (VP1 and VP2). Subsequently, the virus has been detected in children with respiratory tract infection in the world including Europe, Asia, Africa, America, and Australia. Among these studies, the prevalence of HBoV infection has been described to vary considerably between 1.5% and 19% [Kesebir et al., 2006; Ma et al., 2006; Sloots et al., 2006; Smuts and Hardie, 2006; Allander et al., 2007; Bastien et al., 2007; Chieochansin et al., 2007; Allander, 2008; Calvo et al., 2008; Jacques et al., 2008; Smuts et al., 2008; Zeng et al., 2010; Zheng et al., 2010]. Recently, HBoV has been detected in 0.5–9.1% of fecal specimens obtained from children with acute gastroenteritis in Spain, Germany, Hong Kong, Korea, China, Japan, and Brazil [Albuquerque et al., 2007; Lau et al., 2007; Lee et al., 2007; Vicente et al., 2007; Nakanishi et al., 2009].
The present study aimed to screen stool samples collected from Japanese and Thai children with acute gastroenteritis during 2005–2008 for HBoV infection, one of less explored viral pathogens which has been reported to be associated with diarrhea recently, and to characterize the detected HBoV strains.
MATERIALS AND METHODS
Clinical Specimens
A total of 329 fecal specimens which had been shown to be negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus [Chanit et al., 2009; Khamrin et al., 2010] and which were collected from infants and children aged from 2 months to 15 years in pediatric clinics encompassing five localities (Sapporo, Tokyo, Maizuru, Osaka, and Saga) in Japan (247 samples collected from July 2007 to June 2008), Thailand (82 samples collected from January to December 2005) were subjected to screening for HBoV. When HBoV was detected, the corresponding patients were identified and their clinical features, laboratory results, and outcome were analyzed retrospectively.
Detection and Genotyping of the Virus
Nucleic acid was extracted from 140 µl of 10% fecal suspension using the QIAamp viral RNA Mini kit (Qiagen, Inc., GmbH, Hilden, Germany) according to the manufacturer's instructions. This product recovers both RNA and DNA from samples allowing other less-explored enteric pathogens such as Aichi virus and human parechovirus being screened. Samples were then screened by polymerase chain reaction (PCR) for the presence of HBoV by using the primers 188F and 542R to amplify a 354-bp PCR product of the NP-1 region [Allander et al., 2005]. In addition, these samples were screened for Aichi virus and human parechovirus also by RT-PCR with published primer sets [Yamashita et al., 2000; Pham et al., 2010a,b].
For genotyping of the detected virus, all HBoV-positive samples were subjected to VP1/VP2 gene sequencing, in which the primers VP1/VP2F and VP1/VP2R were used to amplify an 819-bp segment of the VP1/VP2 gene [Kesebir et al., 2006].
Sequence Analysis
The PCR amplicons of the NP-1 and VP1/VP2 genes were purified and sequenced in both directions using the BigDye Terminator Cycle Sequencing kit (Perkin Elmer-Applied Biosystems, Inc., Foster City, CA). The primers for amplification of NP1 and VP1/VP2 genes were used as sequencing primers. The sequence data were collected by an ABI Prism 310 Genetic Analyzer (Perkin Elmer-Applied Biosystems, Inc.).
The nucleotide sequences of the NP-1 and VP1/VP2 genes were compared to those of HBoV strains available in GenBank. The sequence data and phylogenesis were analyzed using BioEdit v7.0.5. A parsimony analysis was conducted using MEGA version 3.1 to determine the evolutionary relationship among studied sequences [Kumar et al., 2004].
The nucleotide sequences of the studied HBoV strains described in this study have been deposited in GenBank under accession numbers: GU563333–GU563338 (NP-1 gene) and GU563339–GU563344 (VP1/VP2 gene).
RESULTS
Of the 329 samples tested, 6 (1.8%) were positive for HBoV. Of these, five samples were collected from Japan and one sample was from Thailand. As a result, the detection rates of HBoV in each country were 2% and 1.2%, respectively. For the five Japanese HBoV-positive samples, three were collected in Maizuru and the two remaining were from Osaka.
For clinical characterization, only data from five Japanese patients were available for analysis. All five Japanese children whose stool specimens showed positive for HBoV were infants aged from 8 to 19 months. All positive samples were detected in either April or May. Diarrhea in these patients lasted from 3 to 6 days, and the number of diarrheal episodes per day was <10. None of the patients was noted with fever, vomiting, dehydration, or respiratory symptoms. Co-detections of HBoV and HPeV or Aichi virus were not found (Table I).
| Samples no. | Month of detection | Gender | Age (month) | Diarrhea | Fever | Vomiting | Wheeze, cough, coryza | Dehydration | Co-infection (AiV and HPeV) | Group | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of diarrheal stools/24 hr (time) | Duration of diarrhea (day) | ||||||||||
| 8384 | April | F | 12 | 2 | 4 | — | — | — | — | — | 2 |
| 8387 | April | M | 12 | 10 | 3 | — | — | — | — | — | 1 |
| 8582 | May | M | 19 | 2 | 4 | — | — | — | — | — | 1 |
| 8583 | May | M | 8 | 4 | 4 | — | — | — | — | — | 1 |
| 8588 | May | M | 9 | 6 | 6 | — | — | — | — | — | 1 |
- M, male; F, female.
All positive PCR products of the NP-1 gene were confirmed by sequencing (GenBank). The virus detected in these patients was defined as HBoV by phylogenetic analysis (Fig. 1A). For the classification of the detected viruses, Figure 1B showed that: four Japanese studied HBoV strains were clustered into group 1, while the remaining Japanese strain and a unique Thai strain belonged to group 2.
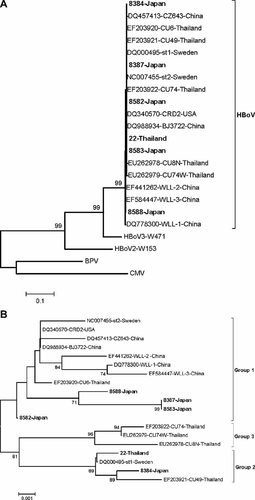
A: Phylogenetic analysis of the NP-1 gene of HBoV in which the other members of Bocavirus genus, human bocavirus 2 (HBoV2), human bocavirus 3 (HBoV3), bovine parvovirus (BPV), and canine minute virus (CMV) were served as the outgroup sequences. B: Phylogenetic tree constructed from 819 nucleotide sequences of the VP1/VP2 gene of the studied and reference HBoV strains. The trees were created by neighbor-joining method and bootstrapped with 500 replicates. Percentage bootstrap values above 70% are shown at the branch nodes. The studied HPoV strains are in bold.
DISCUSSION
In this study, detection rates of HBoV in the samples tested from Japan and Thailand were found to be 2% and 1.2%, respectively. However, they do not represent the percentages of cases of infection with HBoV in all patients with acute gastroenteritis during 1-year period, because only samples which had been known to be negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus were screened for HBoV. As a result, possible mixed infection of enteric viruses was excluded from analysis.
To date, HBoV has been detected in samples from respiratory tract and has been associated with both upper and lower respiratory tract diseases in infants and children [Kesebir et al., 2006; Ma et al., 2006; Sloots et al., 2006; Smuts and Hardie, 2006; Allander et al., 2007; Bastien et al., 2007; Chieochansin et al., 2007; Allander, 2008; Calvo et al., 2008; Jacques et al., 2008; Smuts et al., 2008; Zeng et al., 2010; Zheng et al., 2010]. HBoV was also present in gastrointestinal tract in children with gastroenteritis [Albuquerque et al., 2007; Lau et al., 2007; Lee et al., 2007; Vicente et al., 2007; Nakanishi et al., 2009]. However, its role in patients with acute gastroenteritis is unclear. In this study, HBoV was detected in stool samples collected from infants with acute gastroenteritis without symptoms of respiratory infection. In addition, the absence of any other intestinal pathogens such as rotavirus, adenovirus, norovirus, sapovirus, astrovirus, human parechovirus, and Aichi virus suggests that this virus is an enteric pathogen, a possible causative agent of diarrhea in the studied patients.
In Japan, the detection of HBoV in rectal swabs from children with acute gastroenteritis was reported once in Sapporo in northern Japan [Nakanishi et al., 2009]. In that study, four samples were positive for HBoV by RT-PCR with the detection rate of 0.5%; however, HBoV genotyping was not done in the four samples. In the present study, the virus was found in infants and children with acute gastroenteritis in Maizuru and Osaka located in southern Japan. In Thailand, HBoV detection in children with acute gastroenteritis was first reported in Bangkok (HBoV group 3, detection rate of 0.9%) [Chieochansin et al., 2008]. Taken together with the previous studies, the findings suggest that this virus may be a possible and infrequent causative agent of Japanese and Thai infants and children with acute gastroenteritis.
To our knowledge, this is the first report of the circulating of HBoV groups 1 and 2, and of HBoV group 2 in infants and children with acute gastroenteritis in Japan and Thailand, respectively. The findings provide better understanding on the epidemiological characteristics of HBoV infections in Japan and Thailand and contribute useful data for future research into HBoVs.
Acknowledgements
We thank Dr. Nobuhisa Ishiguro (Department of Pediatrics, Hokkaido University Graduate School of Medicine) for kindly providing a positive control strain of human bocavirus.




