Genetic and physiological characterization of yeast isolated from ripe fruit and analysis of fermentation and brewing potential
Abstract
‘Wild’ and spontaneously fermented beers are growing in popularity in the craft beer industry. Most of these beers are fermented by the use of either pure cultures of unconventional yeast and bacteria or spontaneous fermentation using mixed local microflora. This study examined the potential of using pure strains of new isolates of wild yeast in the fermentation of a unique beer. The microbial communities from the fruit of pindo palm, loquat, hackberry and blackberry were collected in liquid culture, then plated for isolation. Ten isolates were selected for further analysis. Strains were identified by restriction fragment length polymorphism (RFLP) analysis and analysed for growth in a simple liquid media, fermentation in a complex media, alcohol tolerance and acid tolerance. Despite identification of some strains as the same species, they displayed a wide range of physiological properties. All strains were tolerant of pH values as low as 2.4, but none were tolerant of pH 1.9. Alcohol tolerance of different strains varied from 6 to 12%. Several strains had properties that suggest potential as primary fermenters, including the alcohol fermentation of a beer wort. Organoleptic properties of beers fermented with several of the strains demonstrated potential for commercial brewing. Copyright © 2014 The Institute of Brewing & Distilling
Introduction
Domestication of wild Saccharomyces cerevisiae yeast revolutionized commercial fermentation, allowing winemakers and brewers to produce a more consistent and predictable product. There are hundreds of yeast strains available to the brewing industry, each with a unique physiology and fermentation profile. Isolation and characterization of these unique strains of yeast ensures that a wide range of yeast-derived organoleptic characters are available to the brewer. It is standard practice in the modern brewery to inoculate the wort with a pure yeast culture to initiate fermentation. This method ensures a healthy fermentation and production of desired aroma and flavour characteristics in the final product 1-4.
In traditional lambic and Flanders sour ale brewing, spontaneous fermentation is employed, in which brewery-resident microbiota inoculate the cooling wort to initiate the fermentation 5-8. A complex succession of yeast and bacteria over a period of up to three years in oak barrels yields a unique, complex, acidic product 6, 9. Blending of old and young beer is often employed to achieve the desired flavour profile. Yeast strains from the genus Dekkera/Brettanomyces (herein referred to as Brettanomyces) contribute essential organoleptic qualities to these beers 5, 10. Interest in these styles has grown dramatically in recent years in the American craft brewing industry 11; however, only a small number of breweries use open spontaneous fermentation, as this method is unpredictable and may lead to high economic losses to the brewer. Other breweries ferment unique wild ales, using pure isolated strains of yeast and bacteria, most originating from Belgian breweries. Compared with S. cerevisiae, only a small number of pure strains of Brettanomyces and lactic acid bacteria are available to the commercial brewer. Since these yeast species are known to live in a wide range of habitats, we wished to explore the potential of uncharacterized wild yeast for use in commercial brewing. To this end, a variety of autochthonous yeasts were collected from fruit skins, and other sources in different environments, to characterize their potential for use in the brewing industry.
For this study, the microflora from pindo palm fruit (Butia capitata), and loquat fruit (Eriobotrya japonica) from northeast Florida, and hackberry (Celtis sp.) and blackberry fruit (Rubus sp.) from southeastern Pennsylvania, were collected and isolated. In order to function in a brewery setting, yeast strains must be alcohol-tolerant and capable of assimilating and fermenting the sugars found in beer wort. Ten isolates were identified by restriction fragment length polymorphism (RFLP) mapping and characterized for their relevant physiological properties. Strains represented the genera Brettanomyces, Candida and Issatchenkia. Several independent isolates of two species were identified, but all isolates had unique physiological properties. Alcohol tolerance ranged from 6 to >12%, and their fermentation performance was equally variable between isolates. Several strains possessed properties that demonstrated potential for use in the brewing industry.
Materials and methods
Collection, isolation and morphological analysis of strains
To collect microbes, a sterile cotton applicator was dampened in MYPG medium (0.3% malt extract, 0.3% yeast extract, 0.2% peptone, 1% glucose) and used to swab ripe fruit from each source. The swab was then transferred to 3–5 mL of sterile MYPG in culture tubes to release the microbes. The tubes were incubated at 25 °C for up to one week, with microbial growth evident after 2 days. The cultures were streaked onto MYPG agar plates (MYPG supplemented with 1.5% granulated agar). Colony morphology was used to identify possible yeast isolates, confirmed by microscopy. For cell morphology, a sterile needle was used to remove a sample from an isolated colony, and dispersed in 15 μL sterile H2O on a microscope slide. A coverslip was added and cells observed on an Olympus BX-60 microscope at 400× magnification. Cells from selected colonies were picked with a sterile inoculating loop and used to inoculate 5 mL MYPG media. Samples of 1 mL of 3–5 day cultures were stored at −80 °C with 20% glycerol as a cryopreservative. Cultures were streaked onto MYPG agar plates to isolate colonies for further analysis.
PCR/RFLP
Strains were identified to the species level by amplification of the 5.8S rDNA gene and flanking internal transcribed spacer (ITS) region as described 12 with modifications. Briefly, 0.5 μL of stationary culture cells were added to 24.5 μL of sterile H2O containing 0.2 μM ITS1 (5′TCCGTAGGTGAACCTGCGG) and ITS4 (5′TCCTCCGCTTATTGATATGC) primers. Then 25 μL 2× PCR Master Mix (Promega) was added and the sample mixed. Cells were used directly for polymerase chain reaction (PCR) as this method yielded more consistent results than several different DNA extraction methods. PCR conditions were as follows: 95 °C, 5 min, followed by 35 cycles of 94 °C 1 min (denature), 55 °C 2 min (anneal), and 72 °C 2 min (extension). A final 8 min extension at 72 °C followed the last cycle. Product length was determined by electrophoresis of 5 μL of sample on a 1.2% agarose gel in E buffer (40 mM Tris-acetate, 0.1 mM EDTA). Then 12 μL of each PCR product was digested separately with the restriction enzymes HhaI (isochizomer of CfoI), HaeIII and HinfI (Promega) as recommended by the manufacturer. Digestion products were resolved on 2% agarose gels. Banding patterns were compared with published records for species identification 13-15. For two isolates, PCR products were sequenced using the ITS1 and ITS4 primers using a Beckman Coulter CEQ8000 Genetic Analysis System according to the manufacturer.
Physiological properties
The ability of isolates to assimilate and grow on a single carbon source was assessed with API 20C test strip (bioMérieux) using the manufacturer's instructions. The pH tolerance was determined by growth in a 10-fold dilution series of lactic acid in MYPG medium, from 1 to 10μM. These dilutions corresponded to pH values of 1.9, 2.4, 2.9, 3.4, 3.9 and 4.4. MYPG/lactic acid (200 μL) was aliquotted in 96-well microtitre plates in triplicate. Following this, 2 μL of 3 day culture was added to each well, and growth was analysed visually after 4 days. Alcohol tolerance was determined by the addition of absolute ethanol to MYPG, to final concentrations of 0–12%. Water was used as a volume adjustment to keep the nutrient concentration consistent for all samples. Medium was distributed into the wells of a 96-well microtitre plate in triplicate. A 2 μL aliquot of culture was added to each well and growth was assessed visually after 4 days.
Wort attenuation
To assess the beer fermentation potential, three different worts were prepared. Worts 1 and 2 were 10 and 12°P worts, respectively, prepared from Briess light dry malt extract, with an average analysis of 13% glucose, 48% maltose, 14% maltotriose and 19% higher dextrins (analysis from Briess.com). Wort 3 was from a 14°P mash of malted barley without carbohydrate analysis. All densities were determined by refractometer, using a wort correction factor of 1.04. For worts 1 and 3, 50 mL of wort in flasks was inoculated with yeast cells to a density of 1 million cells per millilitre. Flasks were foil covered and incubated with shaking at room temperature for 4 weeks. Wort 2 was from the beer analysis procedure described below. Final brix readings were taken with a refractometer and converted to specific gravity to determine the real and apparent attenuation using a web calculator 16. Not all strains were tested in all three worts.
Beer analysis
Wort (10 L of 12°P) was prepared from Briess light dry malt extract. Bittering hops were added to 30 IBU (calculated). No flavour or aroma hops were added. After cooling and aerating with pure oxygen, 400 mL of wort was added to a 500 mL PET bottle fermenter with an airlock. Starter culture ( 40 mL) was added to each wort sample and fermented at ~20 °C (68 °F) for 4 weeks. Beer was decanted to a 350 mL sanitized bottle, dextrose solution added to generate ~2.5 vol. CO2, and capped. The remaining beer was used to determine the final gravity. After 4 weeks, carbonated beer was chilled and judged for general drinkability by a panel of four experienced beer judges.
Results
Isolation/microscopy/colony morphology
Fruit skins serve as a habitat for a diverse microbial community including yeasts 17. Pindo palm (Butia capitata), loquat (Eriobotrya japonica), blackberry (Rubus sp.) and hackberry (Celtis sp.) fruits were sampled for analysis of resident microbes. Samples were amplified in liquid culture then plated for isolation, and a variety of putative bacterial and yeast colonies were observed. A total of 10 isolates were selected for further analysis based on cell and colony morphology. Isolates were designated with the genus/species initials of the source plant (Bc for pindo palm, Ej for loquat, Rs for blackberry and Cs for hackberry) followed by a numerical identifier.
Colony morphology varied between isolates. All colonies were off-white to cream, and butyrous, dry or rough in surface appearance. Colony shapes were circular to irregular with entire or undulate margins. Elevations ranged from flat to cratered. Specific features of colony morphology for each isolate are described in Table 1. Cell morphology was variable between isolates, within a single isolate, and from a single colony or liquid culture. Samples ranged from primarily single ovoid cells, to highly variable samples with single cells or branched chains, and from ellipsoid to highly elongate. Chains were single or branched, depending on the isolate. Cell morphology is described in Table 1.
| Strain | Colony (MYPG agar) | Cell size (µm) | Cell morphology |
|---|---|---|---|
| Bc01 | Circular; raised or umbonate elevation; entire margin | 2–4 × 4–10 | Ellipsoidal to elongate; single or in short chains |
| Bc02 | Circular to irregular with fine undulate margin; crateriform elevation | 2–6 × 6–15 | Variable; globose to elongate; single with occasional short chains |
| Bc04 | Irregular with high, crateriform elevation; lobate margin | 3–4 × 10–13 | Mostly elongate with large, branched chains |
| Bc07 | Circular to irregular with fine undulate margin; raised elevation | 2–4 × 5–12 | Ellipsoidal to elongate; mostly as single cells |
| Bc08 | Circular to irregular with fine undulate margin; flat to slightly raised elevation | 2–4 × 5–8 | Ovoid; single cells |
| Bc10 | Circular to irregular with fine undulate margin; flat to slightly raised elevation | 2–5 × 4–8 | Ovoid to ellipsoidal; singles, pairs or short chains |
| Bc11 | Circular to irregular with fine undulate margin; crateriform elevation | 2–6 × 6–15 | Mostly ovoid as singles or small clusters; occasional elongate in branched chains |
| Rs01 | Circular; raised elevation; entire margin | 3–4 × 4–8 | Mostly ovoid in singles or small clusters; occasional elongate |
| Cs01 | Circular; raised elevation; entire margin | 2–4 × 4–7 | Mostly ovoid in singles; rare elongate cells |
| Ej02 | Circular; raised elevation; entire margin | 3–4 × 5–7 | Mostly ovoid in singles; rare elongate cells |
RFLP analysis of the 5.8S rDNA and flanking ITS region
In order to determine the identity of the isolates, the 5.8S rDNA and flanking ITS region was amplified by PCR and analysed by electrophoresis to determine amplified product size 12. PCR products were then digested with restriction enzymes HhaI (isoscizomer of CfoI), HaeIII and HinfI. Digestion products were separated on 2% agarose gels, and band patterns were compared with published yeast RFLP databases 13-15. The results are shown in Table 2. Eight of the 10 isolates matched the banding pattern of all three enzymes for Candida incommunis (Bc01), Brettanomyces bruxellensis (isolates Bc02, Bc07, Bc11), Issatchenkia terricola (Bc04) and Brettanomyces anomalus (Rs01, Cs01, Ej02). Banding patterns for Bc08, and Bc10 were identical, but did not match published data. The PCR products were therefore resolved and isolated, followed by sequence analysis using either the ITS1 or ITS4 primers. Sequence data (NCBI GenBank accession number KF921021) was compared with the National Centre for Biotechnology Information non-redundant nucleotide database using standard BLAST parameters. BLAST results confirmed the identity of these isolates as Candida diversa.
| Strain | PCR | Restriction fragments | Identification | Reference | ||
|---|---|---|---|---|---|---|
| HhaI | HaeIII | HinfI | ||||
| Bc01 | 420 | 400 | 420 | 210 + 190 | Candida incommunis | 13 |
| Bc02 | 460 | 230 + 130 + 80 | 330 + 95 + 65 | 265 + 105 | Brettanomyces bruxellensis | 15 |
| Bc04 | 430 | 130 + 100 + 90 | 290 + 130 | 230 + 105 + 105 | Issatchenkia terricola | 13 |
| Bc07 | 460 | 230 + 130 + 80 | 330 + 95 + 65 | 265 + 105 | B. bruxellensis | 15 |
| Bc08 | 430 | 195 + 90 | 295 + 145 | 420 | Candida diversa | This study |
| Bc10 | 420 | 195 + 90 | 295 + 145 | 420 | Candida diversa | This study |
| Bc11 | 460 | 230 + 130 + 80 | 330 + 95 + 65 | 265 + 105 | B. bruxellensis | 15 |
| Rs01 | 800 | 340 + 340 + 120 | 800 | 360 + 190 + 160 + 80 | Brettanomyces anomalus | 15 |
| Cs01 | 800 | 340 + 340 + 120 | 800 | 360 + 190 + 160 + 80 | B. anomalus | 15 |
| Ej02 | 800 | 340 + 340 + 120 | 800 | 360 + 190 + 160 + 80 | B. anomalus | 15 |
Carbon assimilation
API 20C test strips (bioMérieux) were used to determine the assimilation of carbon sources. All isolates could assimilate glucose. All isolates could assimilate glycerol except the three B. anomalus strains and one isolate of B. bruxellensis (Bc11). The three B. bruxellensis isolates were otherwise identical in this test, except for the degree of assimilation of N-acetyl-glucosamine, which was weak and delayed in Bc02 and Bc07 compared with Bc11. Only one other isolate could assimilate N-acetyl-glucosamine, the C. incommunis isolate (Bc01). The I. terricola isolate Bc04 could only assimilate glucose and glycerol of the compounds tested. The two isolates of C. diversa were the only strains able to assimilate D-xylose. The C. incommunis isolate was able to assimilate 13 of 19 compounds, far more than any other strain tested. No isolate was able to assimilate L-arabinose, adonitol, D-lactose, D-melizitose or D-raffinose. Two (C. diversa) or three (B. bruxellensis, B. anomalus) independent isolates of three species were tested. The multiple isolates were very similar within each species, but each isolate had unique properties in these tests. The exception was Ej02, one of the three B. anomalus isolates, which differed considerably from the other two strains. Ej02 was unique among all of the Brettanomyces isolates in its ability to assimilate galactose, maltose and sucrose. It could not assimilate 2-keto-gluconate or cellobiose like the other two B. anomalus isolates. The observed assimilation results generally correlated well with the published data, although there were some differences from the type strains for some of these species 18-22. Assimilation data is summarized in Table 3.
| Substrate | Bc01 | Bc02 | Bc04 | Bc07 | Bc08 | Bc10 | Bc11 | Rs01 | Cs01 | Ej02 |
|---|---|---|---|---|---|---|---|---|---|---|
| D-Glucose | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Glycerol | ++ | + | ++ | + | ++ | ++ | − | − | − | − |
| 2-Keto-gluconate | ++ | − | − | − | − | + | − | ++ | ++ | − |
| L-Arabinose | − | − | − | − | − | − | − | − | − | − |
| D-Xylose | − | − | − | − | ++ | ++ | − | − | − | − |
| Adonitol | − | − | − | − | − | − | − | − | − | − |
| Xylitol | ++ | − | − | − | − | ++ | − | − | − | − |
| D-Galactose | + | − | − | − | − | − | − | − | − | ++ |
| Inositol | ++ | − | − | − | − | − | − | − | − | − |
| D-Sorbitol | ++ | − | − | − | − | − | − | − | − | − |
| Methyl α-D-glucopyranoside | ++ | − | − | − | − | − | − | − | − | − |
| N-Acetyl-glucosamine | ++ | + | − | + | − | − | ++ | − | − | − |
| D-Cellobiose | ++ | − | − | − | − | − | − | ++ | ++ | − |
| D-Lactose | − | − | − | − | − | − | − | − | − | − |
| D-Maltose | ++ | − | − | − | − | − | − | − | − | ++ |
| D-Sucrose | ++ | − | − | − | − | − | − | − | − | ++ |
| D-Trehalose | ++ | − | − | − | − | − | − | − | − | − |
| D-Melezitose | − | − | − | − | − | − | − | − | − | − |
| D-Raffinose | − | − | − | − | − | − | − | − | − | − |
- a −, No growth; +, weak, delayed growth; ++, growth.
pH and alcohol tolerance
The strain's limits for growth in an acidic or alcoholic environment were tested. The pH of MYPG medium was adjusted by the addition of lactic acid to 1.0 M, followed by serial 10-fold dilutions. Overnight culture (2 μL) of each isolate was inoculated into 200 μL of pH-adjusted MYPG in wells of a 96-well microtitre plates in triplicate. After 4 days at 25 °C, growth was assessed visually, and the results are shown in Table 4. Bc01 showed limited growth at pH 2.4, while all of the other isolates grew well at this pH. No strain grew at the most acidic pH of 1.9. Similarly, growth was assessed in ethanol ranging from zero to 12% (vol%). Significant variation between strains was observed for alcohol tolerance. The C. incommunis isolate was the least tolerant. It grew well at 4%, but was nearly completely inhibited at 6%. The two C. diversa isolates were similar to one another, growing poorly at 8%, and were completely inhibited above this concentration. Bc10 was similar to Bc08, except that it grew robustly at 0–4% compared with Bc08. Both strains were inhibited at 6 and 8%, and did not grow at 10%. I. terricola required 12% ethanol to completely inhibit growth, although growth was slower at 8 and 10%. All of the B. bruxellensis and B. anomalus isolates grew in 12%, the highest ethanol concentration tested. Isolates Bc02 and Bc07 grew more robustly at zero and 2% ethanol than at higher concentrations, but there was no further reduction in growth through to 12%. The Bc11 isolate showed no growth inhibition at any ethanol concentration, as did the Rs01, Cs01 and Ej02 strains. Data for growth in ethanol is shown in Table 5.
| pH | Bc01 | Bc02 | Bc04 | Bc07 | Bc08 | Bc10 | Bc11 | Rs01 | Cs01 | Ej02 |
|---|---|---|---|---|---|---|---|---|---|---|
| No yeast | − | − | − | − | − | − | − | − | − | − |
| 5.0 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 4.4 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 3.9 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 3.4 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 2.9 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 2.4 | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 1.9 | − | − | − | − | − | − | − | − | − | − |
- a −, No growth; +, minimal growth; +++, robust growth.
| Alcohol | Bc01 | Bc02 | Bc04 | Bc07 | Bc08 | Bc10 | Bc11 | Rs01 | Cs01 | Ej02 |
|---|---|---|---|---|---|---|---|---|---|---|
| No yeast | − | − | − | − | − | − | − | − | − | − |
| 0.0% | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 2.0% | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ |
| 4.0% | +++ | ++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| 6.0% | + | ++ | +++ | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| 8.0% | − | ++ | ++ | ++ | + | + | +++ | +++ | +++ | +++ |
| 10.0% | − | ++ | + | ++ | − | − | +++ | +++ | +++ | +++ |
| 12.0% | − | ++ | − | ++ | − | − | +++ | +++ | +++ | +++ |
- a −, No growth; +, minimal growth; ++, moderate growth; +++, robust growth.
Attenuation and beer analysis
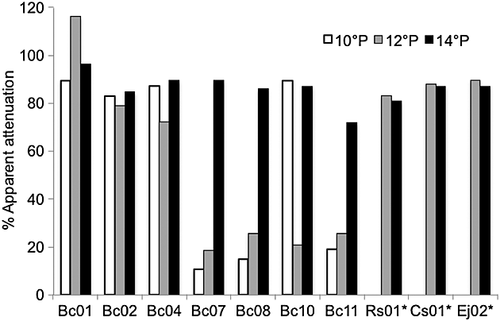
In order to be viable as a brewing strain, yeast must be able to carry out ethanol fermentation of simple sugars. To test fermentation potential, strains were inoculated into beer wort. Three different worts (10, 12 and 14°P) were inoculated with log phase cells and incubated 28 days at approximately 20 °C. Final gravity readings were measured and the real and apparent attenuation was determined (Fig. 1). Owing to the timing of yeast collection, not all strains were tested in all three worts. All of the isolates attenuated the 14°P wort to at least 70% apparent attenuation; however, the strains were highly variable in their attenuation of the remaining wort samples. The C. incommunis isolate (Bc01), B. bruxellensis isolate Bc02, I. terricola isolate and all three B. anomalus isolates fermented all wort samples to an apparent attenuation >70%. Three of the isolates (B. bruxellensis isolates Bc07 and Bc11, and C. diversa isolate Bc08) were poor fermenters on the other two worts, achieving attenuation of only 20–25%. The other C. diversa (Bc10) isolate fermented two samples to nearly 90% apparent attenuation, but only 20% on the remaining wort. It is notable that different isolates of the same species (C. diversa and B. bruxellensis) showed widely variable results in this assay. Only the B. anomalus and C. incommunis isolates were consistent and efficient fermenters.
Beer fermented with each strain was analysed by a panel of experienced competition beer judges. Analysis was restricted to fermentation character. Most strains exhibited a strong phenolic character of plastic, medicinal and burnt or smoky. The beer fermented by strains that had poor attenuation of this wort exhibited a sweet, under-attenuated worty character. Several of the strains produced beer with a more desirable complex, spicy and fruity character. Owing to the subjective nature of this analysis, beers were categorized as ‘unlikely to succeed as a brewing strain’, or ‘shows some brewing potential’. While somewhat variable in this single sample, all three B. anomalus strains were characterized as having brewing potential, while none of the other strains met this criteria.
Discussion
Yeasts are ubiquitous organisms. A wide variety of genera can be found on fruit skins, grains and other environments related to commercial beverage fermentation. Ten yeast isolates from several ripe fruit sources were identified and characterized. Several isolates were determined to be the same species based on RFLP mapping or DNA sequencing of the 5.8S rDNA region, but all showed unique physiological characteristics. Between species, there was significant variation in physiology, particularly with regards to ethanol tolerance and the fermentation of complex media (wort). Isolates of B. bruxellensis, I. terricola, C. diversa and B. anomalus had properties appropriate for fermentation, in particular efficient attenuation of beer wort, and an alcohol tolerance to ≥8%. Three other strains were deficient in fermentation, achieving an apparent attenuation of 25% or less in several experiments. The C. incommunis isolate was the most efficient fermenter, achieving an apparent attenuation consistently over 90%. Interestingly, this was also the strain with the lowest alcohol tolerance, showing reduced growth at 6% alcohol and complete inhibition at 8%. The worts tested in the experiments would yield alcohol to about 7% if fully fermented, within the tolerance range determined for this strain. This strain would be expected show a lower percentage attenuation of worts with significantly higher original gravity, as the higher alcohol yield would inhibit growth.
It has been observed that metabolic properties and the yeast-derived compounds present in the finished beer vary when Brettanomyces strains are used as the sole fermentative organism vs their use as part of a mixed fermentation 23. There is further variation based on whether a mixed fermentation contains only Saccharomyces yeast in addition to Brettanomyces, or whether lactic acid- or acetic acid-producing bacteria are also present 6, 10, 24. It has been proposed that Brettanomyces generate more organoleptic complexity when in a mixed fermentation as a result of their need to compete for limited resources with the more robust Saccharomyces strains 25. A higher acid content creates more substrates for ester formation by Brettanomyces, adding additional complexity to the finished product 24. If the beer is packaged with Brettanomyces for bottle-conditioning, the long-term storage results in a continual evolution of the aroma and flavour characteristics of the beer. Further analysis of the B. anomalus isolates described here under these variable fermentation conditions is warranted.
It is well recognized that yeast are abundant in nearly every environment; however, only a minority from any particular source are likely to possess physiological characteristics for potential use in commercial fermentations. The sugar composition and other nutrient resources of fruit and beer wort are distinct, requiring efficient adaptation by these yeasts to the new environment. Even fewer strains will exhibit desirable organoleptic properties. A subjective analysis of the fermentation characteristics of beer produced by new strains described here determined that all three B. anomalus isolates expressed acceptable properties with no overt off-flavours or aromas. The Ej02 B. anomalus strain isolated from loquat was used to ferment 100 L of an 800 L recipe of saison-style ale brewed on an 800 L commercial system. Full 800 L fermentations are planned once appropriate pitching volumes of yeast are generated. While long-term adaptation to a brewery environment may eventually lead a mixed wild population towards desirable properties, this process is uncertain and financially untenable to most commercial breweries. Isolation and characterization of new strains as described here may overcome these obstacles and offer new strain options to adventurous brewers.
Acknowledgements
This work was partially supported by an AHA Research and Education Fund grant from the Brewers Association. Trained competition beer judges participated in beer analysis.




