Regulatory effect of NK homeobox 1 (NKX2.1) on melanocortin 4 receptor (Mc4r) promoter in Mandarin fish
Abstract
The melanocortin 4 receptor (MC4R) is a G protein-coupled transporter that mediates the regulation of thyroid hormones and leptin on energy balance and food intake. However, the mechanisms of transcriptional regulation of Mc4r by thyroid hormone and leptin in fish have been rarely reported. The messenger RNA expression of Mc4r gene was significantly higher in brain than those in other tissues of mandarin fish. We analyzed the structure and function of a 2029 bp sequence of Mc4r promoter. Meanwhile, overexpression of NKX2.1 and incubation with leptin significantly increased Mc4r promoter activity, but triiodothyronine showed the opposite effect. In addition, mutations in the NKX2.1 binding site abolished not only the activation of Mc4r promoter activity by leptin but also the inhibitory effect of thyroid hormones on Mc4r promoter activity. In summary, these results suggested that thyroid hormones and leptin might regulate the transcriptional expression of Mc4r through NKX2.1.
Abbreviations
-
- AgRP
-
- Agouti related protein
-
- AMPK
-
- AMP-activated protein kinase
-
- ATCH
-
- adrenocorticotropic hormone
-
- cAMP
-
- cyclic adenosine monophosphate
-
- CCK
-
- cholecystokinin
-
- EGR1
-
- early growth response factor 1
-
- GPCR
-
- G protein-coupled receptors
-
- Mc4r
-
- melanocortin receptor-4
-
- MSH
-
- α-melanocyte-stimulating hormone
-
- NKX2.1
-
- thyroid transcription factor-1
-
- POMC
-
- pro-opiomelanocortin
-
- rpl13a
-
- ribosomal protein L13a
-
- TFBS
-
- transcriptional factor binding sites
-
- TH
-
- thyroid hormones
-
- TRE
-
- thyroid hormone response elements
-
- TSS
-
- transcription start sites
1 INTRODUCTION
Organisms regulate the body's food intake and energy homeostasis via the central and peripheral neural systems to maintain healthy, normal growth, breeding, and other physiological processes (Krashes et al., 2016). Short-term relatively drastic fluctuations in energy homeostasis can be tolerated and corrected, while long-term chronic instability may damage immunity and metabolism (Guh et al., 2009; Olshansky et al., 2005). Melanocortin-4 receptor (MC4R) is a peptide secreted by the ventral medial nucleus of the hypothalamus (Yeo et al., 2021), which plays an important role in the maintenance of energy homeostasis in the organism. MC4R increases intracellular cAMP levels by binding to endogenous agonists such as α-MSH and ACTH secreted by POMC neurons, thereby suppressing appetite, reducing food intake, and increasing energy expenditure. Moreover, MC4R also binds to MC4R antagonists (AgRP) released by AgRP neurons, which limits or reverses the effects of Mc4r basal activity, leading to increased food intake and reduced energy expenditure (Bouret, 2022; Bruschetta et al., 2020; Dores & Baron, 2011; Kleinau et al., 2020).
Individual obesity is often produced by Mc4r mutations in animals (Gonçalves et al., 2018). In fish, the MC4R signaling system plays a vital role in food intake and growth (Zhang et al., 2020). Intraventricular injection of MC4R agonist reduces food intake in zebrafish (Danio rerio) (Josep Agulleiro et al., 2013) and coho salmon (Oncorhynchus kisutch) (White et al., 2016). MC4R has also been implicated to the regulation of fish development, with Mc4r mutants developing considerably better after Mc4r was knocked out using CRISPR/Cas9 in catfish (Ictalurus punctatus) (Coogan et al., 2022). In medaka, Mc4r hinders embryonic development and delays hatching after the gene is knocked out (Liu et al., 2019). Nonsynonymous mutations of Mc4r in cavefish contribute to increased appetite, growth, and hunger resistance (Aspiras et al., 2015).
Transcriptional regulation of genes confers the competence of organisms to respond to intracellular and extracellular signals, and is essential for the physiological function of the organism (Core et al., 2014; Cramer, 2019). The promoter is a critical region of gene expression that contains many functional DNA sequences that interact with regulatory proteins such as transcription factors (TF) and gene-specific DNA binding proteins to integrate information from many signaling pathways (Riethoven, 2010) and control the initiation and intensity of transcription (Mikhaylichenko et al., 2018) thereby regulating gene expression (FitzGerald et al., 2004). Currently, research on Mc4r mainly focuses on the affinity of Mc4r with ligands, so as to explore activators that can precisely target the treatment of obesity (Gonçalves et al., 2018), or to explore the association between Mc4r polymorphisms and obesity (Fatima et al., 2022). However, information on the functional interpretation and transcriptional regulation of the Mc4r promoter in fish is scarce and requires more research.
NK homeobox 1 (Nkx2.1), also known as thyroid transcription factor-1 (TTF-1), is expressed in the mammalian hypothalamus (Gehring et al., 1994) and activates arcuate Pomc expression in mice from early development to adulthood by interacting with the upstream distal enhancers nPE1 and nPE2 motifs of the anorexia neuropeptide POMC (Orquera et al., 2019). Nkx2.1 is also engaged in feeding and trophic homeostasis regulating and operate as a TFs for appetite regulators, such as neuropeptide Y (NPY) and agouti-associated protein (AgRP), among others (Vinnicombe & Volkoff, 2022). As one of the functional receptors of POMC, Mc4r is involved in appetite regulation, and NKX2.1 is expected to contribute in energy balance regulation and appetite regulation through modulating Mc4r transcriptional expression.
Thyroid hormones usually regulate food intake in mammals via the arcuate nucleus of the hypothalamus (ARC) or mediate energy metabolic balance via the central system (Varela et al., 2012). Hyperthyroidism is commonly characterized by hypereating and weight loss (López et al., 2013; Silva, 2006). Transcriptional regulation of Mc4r by thyroid hormone receptor has been reported in obese rats, and different receptors have distinct transcriptional regulatory mechanisms for Mc4r (Decherf et al., 2010; Tabachnik et al., 2017). However, transcriptional regulation of Mc4r by TH has rarely been reported in fish. Leptin is a multifunctional hormone that regulates food intake, body weight, and stress response of organism mainly through the hypothalamic melanocortin system and Mc4r-expressing neurons (Gorissen & Flik, 2014; Jéquier, 2002). The number of molecular studies on fish feeding regulation continues to grow (Clément et al., 2018; Volkoff, 2016), but the regulatory relationship between leptin signaling and the transcriptional activity of the Mc4r gene is unclear.
Mandarin fish is a typical carnivorous fish, a valuable freshwater fish in China, with a unique feeding habit, feeding on live bait for life from the beginning of feeding and refusing to eat dead bait (Yi et al., 2013). The transcriptional regulation mechanism of Mc4r by T3 and leptin is poorly studied, particularly in carnivorous fish, and the gene regulation associated to the special feeding habit and energy metabolism of Mandarin fish needs further investigation. We analyzed the structural function of the Mc4r promoter in Mandarin fish, identified binding site NKX2.1 binding site in its promoter region, and elucidated the regulatory mechanism of Mc4r transcription by NKX2.1, TH, and leptin, providing new insights into the regulation of diet and energy balance in vertebrates.
2 MATERIALS AND METHODS
2.1 Experimental animals and reagents
The Mandarin fish used for DNA and RNA extraction was acquired from the Mandarin Fish Research Center of Huazhong Agricultural University. HEK293T cell lines were obtained from the Cell Resource Center of Fishery College of HZAU. Dulbecco's modified Eagle's medium (DMEM), opti-MEM™, fetal bovine serum (FBS), and 0.25% trypsin-EDTA were purchased from Gibco. Recombinant Mandarin fish leptin was obtained from our laboratory (Yuan et al., 2020), and triiodothyronine purchase (T3; CAS 6893-02-3) (purity ≥95%) was purchased from Sigma-Aldrich.
2.2 Quantitative polymerase chain reaction (qPCR) analysis
Using a multifunction microplate reader, the amount of RNA was measured (Biotek). Following the manufacturer's instructions, 1 μg of total RNA was reverse-transcribed using the HiScript Q RT SuperMix for qPCR (+gDNA wiper) kit from Vazyme. The PCR products were detected using 1.2% agarose gel electrophoresis, which was carried out for 20 min at 140 V. The Gel Imaging System was then used to take pictures of the gel under ultraviolet light. In the following investigations, real-time PCR was used to analyse the level of gene expression. Table 1 contained details regarding relative gene primers. Rpl13a was employed as a housekeeping gene for Mandarin fish (Lu et al., 2023). The AceQ® qPCR SYBR Green-Master Mix kit was used for the qPCR (Vazyme). The Ct value method was used to calculate the relative gene expression (Livak & Schmittgen, 2001).
| Gene | Sequence 5′−3′ | Tm (°C) |
|---|---|---|
| rpl13a | CACCCTATGACAAGAGGAAGC | 59.0 |
| TGTGCCAGACGCCCAAG | ||
| Mc4r | TTGGGGTGTTCGTTGT | 58.0 |
| GGAATGCACAGGCTTG |
- Abbreviation: PCR, polymerase chain reaction.
2.3 Cloning and functional analysis of Mc4r promoter
2.3.1 Sequence analysis and cloning of the promoter region of Mc4r gene
The promoter sequence of the Mc4r gene was obtained from the mandarin fish genome (PRJNA513951) uploaded to NCBI in this experiment (He et al., 2020). Transcription start sites (TSS) were predicted using Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/) (Prestridge, 1995). The protocols of promoter cloning following those in Xu et al. (2017). The promoter sequences prediction was carried out using Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/promoter.html) (Reese, 2001) and the MethPrime Cpgplot (http://www.urogene.org/methprimer/) was used to predict the CpG Island (Choi et al., 2003). Moreover, the TF binding sites were analyzed with ALGGEN–PROMO (https://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/) (Loots et al., 2002; Ovcharenko et al., 2004) and JASPAR (https://jaspar.genereg.net/) (Chen et al., 2020; Zhuo et al., 2020). According to the predicted core region of potential TFs, the regions of the Mc4r promoter from −1708 to +321 bp, −908 to +321 bp, −282 to +321 bp, and −41 to +321 bp were cloned by step deletion method. when the TF NKX2.1 is amplified, the HA-tag protein tag sequence is added at the C-terminus. The primer sequences for amplifying promoters in different regions and NKX2.1 are shown in Table 2.
| Genes | Primers | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| Mc4r | Outer primer (TSS) | GGTTGACATTCGTGGTTT | CAACAGCGAGGCTACAGA |
| Inner primer (TSS) | |||
| −362/+321 | ctatcgataggtaccgagctcTGCTCGTACCTTTCTGCCTT | acttagatcgcagatctcgagTCCCTGTCTTTTTGGGTTAAGTCT | |
| −603/+321 | ctatcgataggtaccgagctcTTACCAGCGAGCCAATAGGAC | acttagatcgcagatctcgagTCCCTGTCTTTTTGGGTTAAGTCT | |
| −1299/+321 | ctatcgataggtaccgagctcGCAACAACTGCAATCAAGCG | acttagatcgcagatctcgagTCCCTGTCTTTTTGGGTTAAGTCT | |
| −1708/+321 | atttctctatcgataggtaccTCAGGATAAATTGGAATCACTTTGG | acttagatcgcagatctcgagTCCCTGTCTTTTTGGGTTAAGTCT | |
| NKX2.1 | Outer primer | CCGCAGGTCTCTCAGGCGGCGAT | TCAAGCGTAATCTGGAACATCGTATGGGTACCACGTCCTGCCGTA |
| Inner primer | ctagcgtttaaacttaagcttATGCAGCAGCACCACATGG | aacgggccctctagactcgagTCAAGCGTAATCTGGAACATCG | |
2.3.2 Plasmid construction
Different regions of the Mc4r promoter were cloned to pGl3-Basic vectors by KpnI and XhoI restriction enzyme sites (Promega). NKX2.1 was subcloned into pcDNA3.1(+) vectors by HindIII and XhoI restriction enzymes. Use the clone express II One Step Cloning Kit (Vazyme) to recombine all recombinants. The constructed fluorescent expression vectors were named as pGl3-41/+321, pGl3-282/+321, pGl3-908/+321, and pGl3-1708/+321, respectively. The overexpression vector was named pcDNA3.1(+)-NKX2.1.
2.3.3 Plasmid transfections and assays of luciferase activities
Human embryonic kidney cells (HEK293) were cultured using DMEM medium containing 10% FBS in a biochemical incubator at 37°C and 5% CO2. In Opti-MEM (Invitrogen) medium, the internal reporter plasmid pRL-TK (20 ng/well) was cotransfected with the plasmid into HEK293 cells by transfection reagent Lipofectamine 2000 (Invitrogen). After 4 h of transfection, change the medium to DMEM containing 10% FBS, incubate for 24 h, and collect samples. Relative luciferase activity was analyzed by the Double Luciferase Reporter Analysis System (Promega).
2.3.4 Protein expression of mandarin fish NKX2.1 in HEK293T cells
The western blot method was used to detect the protein expression of the constructed mandarin fish NKX2.1 protein and the control pcDNA3.1(+) in HEK293T cells. The antibodies selected for this experiment are: β-actin (1:10,000, ABL1011; Abbkine), anti-HA tag (1:5000; Mabnus).
2.3.5 Site-mutation analysis of the NKX2.1 binding sites on Mc4r promoters
To identify the NKX2.1 binding sites on mandarin fish Mc4r promoters, we conducted the site-mutation analysis via the QuickChange II Site-Directed Mutagenesis Kit (Vazyme) according to the manufacturer's instructions. The site-mutation primers are listed in Table 3. Their mutant plasmids were named Mut-Mc4r-NKX2.1. This constructs and pRL-TK were then cotransfected into HEK293T cells via the Lipofectamine 2000 (Invitrogen). After 4 h transfection, the medium was replaced by the DMEM with 10% FBS. After 24 h incubation, the cells were lysed and harvested for the luciferase activities according to the procedure above.
| Gene | Primers | Forward primer (5′−3′) | Reverse primer (5′−3′) |
|---|---|---|---|
| Mc4r | Mut- Mc4r - NKX2.1-1 | TTTAgtagacgtgcggcGACCTCACCCTACCTTTATCATTCA | gccgcacgtctacTAAATAGTATTATTCCTATTATTATTAATATATTTTAAAGTT |
| Mut- Mc4r - NKX2.1-2 | ATcttcagctcgcgtAGTATGTATATTAGTATGTATATAGTATGAAAACTTTAAAA | CTacgcgagctgaagATTACATTTATTTACAATATTTATTTAGTATTTTTATACT |
2.4 Leptin and thyroid hormones regulate the transcription of Mc4r via NKX2.1
The Mc4r promoter plasmid is transfected into cells and then incubated with T3 with concentration gradients of 0, 1, 10, 100, and 500 nM, respectively. The optimal concentration of T3 was screened by the effect of T3 on the activity of the Mc4r promoter. After transfection of Mc4r promoter 5′-deletion fragment plasmids (pGl3-41/+321, pGl3-282/+321, pGl3-908/+321, and pGl3-1708/+321), pcDNA3.1(+)-NKX2.1, and Mut-Mc4r-NKX2.1, the transfected HEK293T cells were incubated using 0 and 200 ng/mL leptin (Lv et al., 2020), and optimal concentrations of T3, respectively. Collect cell samples after 24 h, the cells were lysed and harvested for the luciferase activities according to the procedure above.
2.5 Data analysis
Statistical analysis was performed with SPSS 25.0 software. All data were presented as mean ± SEM (standard error of the mean). The effects of treatment were assessed using one-way analysis of variance after Levene's test for homogeneity of variance, and differences between group means were compared by post hoc Duncan's test. Differences with p < 0.05 were considered to be significant.
3 RESULTS
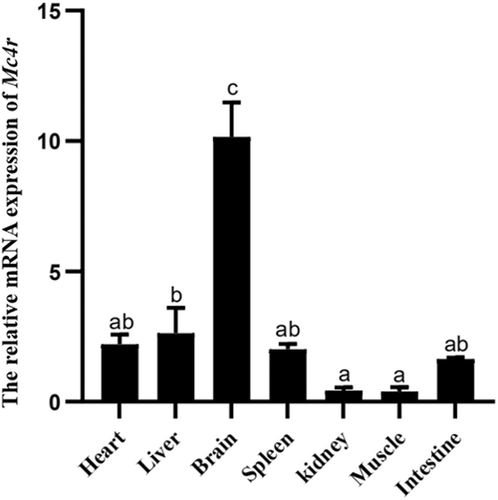
3.1 Tissue distribution of Mc4r gene in Mandarin fish
The Mc4r gene was strongly expressed in the brain, with moderate expression levels in the liver, as determined by real-time PCR (Figure 1).
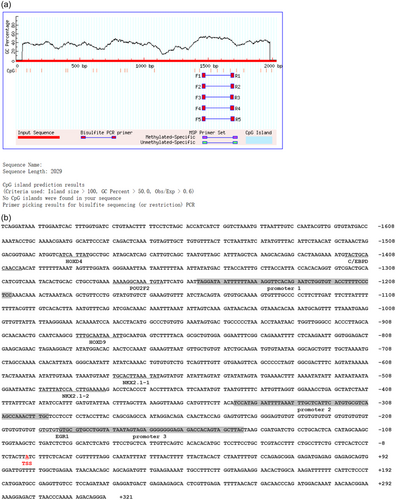
3.2 Cloning and sequence analysis of Mc4r promoters
Mc4r promoter sequences were obtained from mandarin fish genomes uploaded to NCBI (PRJNA513951) (He et al., 2020), and 2029 bp sequences of Mc4r promoters were cloned. Transcription start sites (TSS) were predicted using Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/) (Prestridge, 1995). The online software predicted that there would be no CPG islands were found in the Mc4r promoter region (Figure 2a), with three potential promoter core regions, with an EGR1 binding site (−197/−184 bp), one HOXD9 binding site (−887/−879 bp), One HOXD4 binding site (−1497/−1484 bp), one POU2F2 binding site (−1277/−1264 bp), one C/EBPD binding site (−1414/−1412 bp) and two NKX2.1 binding sites, named NKX2.1-1 (−577/−565 bp) and NKX2.1-2 (−491/−479 bp), respectively (Figure 2b).
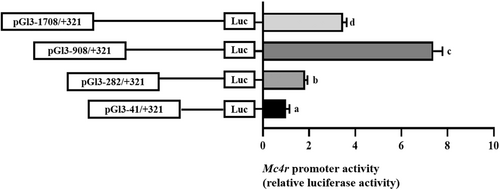
3.3 The relative luciferase activity of 5′-deletion assay of the regions of Mc4r promoters
The unidirectional deletion sequences from −1708 to −908 bp of the Mc4r promoter significantly increased the relative luciferase activity, but the unidirectional deletion sequences from −908 to −41 bp significantly decreased the luciferase activity of the Mc4r promoter, and the results showed that the core region of the Mc4r promoter might exist from −908 to −41 bp (Figure 3).
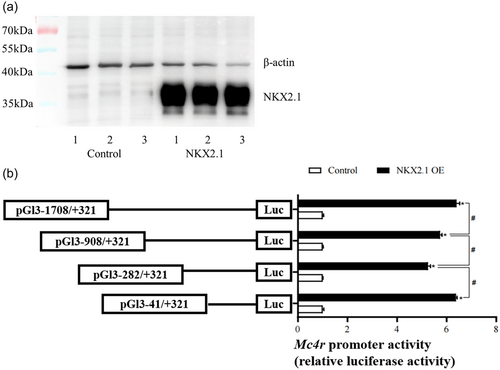
3.4 Overexpression analysis of NKX2.1 in Mc4r promoter region of Mandarin fish in HEK293T cells
After transfecting pcDNA3.1(+)-NKX2.1 plasmid into HEK293T cells for 24 h, it was found to be successfully overexpressed in HEK293T cells by western blot (Figure 4a). Then, pcDNA3.1(+)-NKX2.1 and Mc4r promoter plasmids were cotransfected into HEK293T cells for 24 h. The analysis of relative luciferase activity showed that overexpression of NKX2.1 significantly increased the activity of Mc4r promoter compared with the control group (Figure 4b).
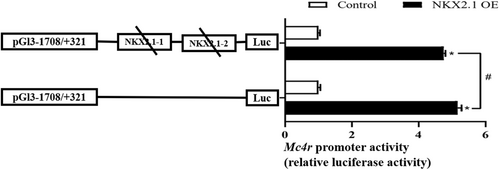
3.5 Site-mutation analysis of NKX2.1 in the Mc4r promoter
To further validate the binding site of NKX2.1 in the Mc4r promoter region and its regulation mechanism on Mc4r promoter activity, the potential binding sites of NKX2.1 (NKX2.1-1 and NKX2.1-2) in the Mc4r promoter region were double-mutated. The activity of the Mc4r promoter was significantly enhanced after overexpression of NKX2.1 compared to negative controls, but when the site was mutated, the degree of activity of the Mc4r promoter was significantly reduced compared to that before the mutation (Figure 5).
3.6 Mc4r promoter transcription responds to thyroid hormones and leptin regulation via NKX2.1
The activity of the Mc4r promoter is dose-dependent on T3 negative feedback regulation when the concentration is between 1, 10, and 100 nM, but its inhibitory effect is diminished when the concentration reaches 500 nM. Thus, 100 nM T3 was adopted in the current study (Figure 6a). To study the responsiveness of the Mc4r promoter to thyroxine and leptin, HEK293T cells were incubated with 100 nM T3 or 200 ng/mL leptin (Lv et al., 2020) for 24 h.
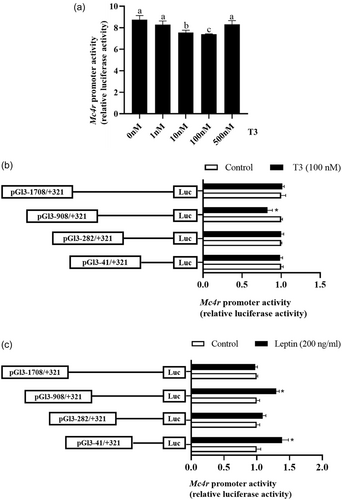
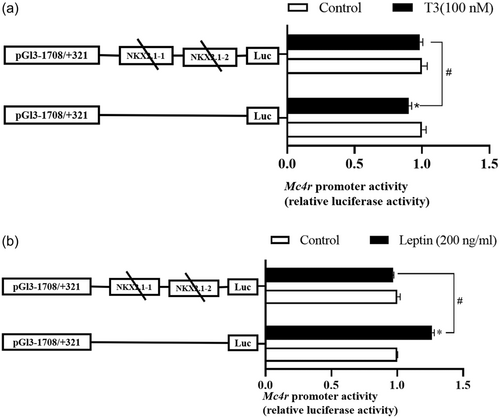
Compared with the control group, T3 significantly inhibited the activity of the Mc4r promoter sequence from −908 to −282 bp, and the activity of the remaining fragments tended to be inhibited, although not significantly (Figure 6b). Meanwhile, leptin incubation increased the relative luciferase activity of the Mc4r promoter compared to the control group, in particular, the relative luciferase activity of the region from −908 to +321 bp and −41 to +321 bp was significantly increased (p < 0.05) (Figure 6c). When compared to the control group, the mutation of the NKX2.1 site of the Mc4r promoter completely counteracts the inhibitory effect of T3 on luciferase activity and improves the activity of the Mc4r promoter (Figure 7a). Furthermore, the activation effect of leptin on Mc4r promoter activity was significantly reduced and virtually completely reversed (Figure 7b). This result implies that NKX2.1 may mediate the transcriptional regulation of Mc4r by T3 and leptin.
4 DISCUSSION
Mc4r is mainly involved in energy balance and appetite regulation in the system, and thyroxine and leptin also play an essential role in the regulation of this physiological process (Kühnen et al., 2019; Vella et al., 2011), while the related molecular mechanisms of transcriptional regulation are less researched. In this study, the Mc4r promoter sequence of mandarin fish was identified for the first time, and a functional TF NKX2.1 binding site in the Mc4r promoter region was discovered to respond directly to thyroxine and leptin. This study contributes to a better understanding of how TH and leptin regulate food intake and energy balance through Mc4r in vertebrates.
In this experiment, the Mc4r gene was expressed in all of the mandarin fish tissues, with brain expression being substantially higher than peripheral tissues. Similar to mammals, Mc4r may be primarily involved in brain pathway regulation (Barb et al., 2010). CpG methylation in the intragenic is a probable factor in regulating gene expression, and higher methylation of CpGs in gene promoters or exons is linked to transcriptional suppression of this gene (Papin et al., 2021; Sarda et al., 2017). The Mc4r promoter region of mandarin fish was predicted, and no CpG islands were detected, indicating that there was no methylation modification in the transcriptional process of Mc4r in mandarin fish. A similar situation has been observed in animals, where no methylation sites were discovered in the Mc4r promoter region of mouse (Tabachnik et al., 2017).
The analysis of promoter core regions is also an important part of the study of promoter transcriptional regulation mechanism (Molloy, 2000), and the promoter core region of the Mc4r gene in humans (Lubrano-Berthelier et al., 2003) and pigs (Zhang et al., 2012) was analyzed separately, and sequence fragments that significantly affected the promoter activity of the target gene were identified. Important regulatory regions were also found in the Mc4r promoter region in mandarin fish, and the Mc4r promoter −1708 to −282 bp greatly influenced the Mc4r promoter activity. Mc4r promoter activity was significantly enhanced when −1708 to −908 bp was consistently absent, indicating that some critical trans-acting factor binding sites that adversely affects Mc4r transcriptional expression activity may present in this region. When the sequence the −908 to −41 bp of the Mc4r promoter was continuously absent in one direction, the promoter activity decreased significantly, indicating that some key positive regulatory elements may be present in this region.
At present, in mammals, the regulatory mechanism of NKX2.1 on POMC is not consistent. It has been reported that NKX2.1 is essential for early formation of hypothalamic melanocortin neuron identity and for participates in the maintenance of Pomc expression levels during adulthood (Orquera et al., 2019). However, higher expression of hypothalamic Pomc was reported in rats knocked down by TTF-1, resulting to anorexia and decreased eating. Yet, infusion of Mc4r antagonists mitigates the decreased feeding caused by TTF-1 antisense oligonucleotide (Kim et al., 2011). Mc4r acts as a functional receptor for POMC (Zhang et al., 2012), and its promoter region predicts two NKX2.1 binding sites in mandarin fish. Overexpression of NKX2.1 significantly increased Mc4r transcriptional activity and the activation effect is significantly reduced when this site is mutated. These results indicate that NKX2.1 can contribute to maintaining energy balance and appetite regulation via regulating the transcription of Mc4r in mandarin fish. When the NKX2.1 TF binding site was mutated, overexpression of NKX2.1 still had an activating effect on MC4R compared to the negative control, probably because this promoter region still has other potential binding sites. The difference between the results of this experiment and those of mammals might be attributed to the fact that NKX2.1 has multiple copies in various species, and there are certain differences in functions between copies. In fish, Nkx2.1 in rainbow trout has four copies due to genome-wide replication, among which Nkx2.1a and Nkx2.1c were found to be unique differentiation types of teleost fish, and each copy had different effects on the transcriptional activity of the promoter, and the luciferase intensity showed that Nkx2.1a significantly stimulated promoter transcription, comparable to Nkx2.1b, while Nkx2.1c only induced weak transcription and had different functions (Suzuki et al., 2005). In mandarin fish, Nkx2.1 contains three transcripts, and the most conserved transcript was selected to construct an overexpression vector.
The activity of the Mc4r promoter decreases as T3 concentrations increase, which is consistent with the negative regulation pattern observed in mammals (Tabachnik et al., 2017). Incubating HEK293T cells with 200 ng/mL leptin enhanced Mc4r promoter activity, which is consistent with positive regulatory tendency in mammals (Caron et al., 2018; Tabachnik et al., 2017). T3 medication significantly reduced the activity of the −282 to −908 bp region of the Mc4r promoter, whereas leptin medication significantly enhanced the activity of this region. Interestingly, two putative binding sites for NKX2.1 were predicted in the −282 to −908 bp region of the Mc4r promoter, and when these two sites were double mutated, both the negative regulation of the Mc4r promoter by T3 and the positive regulation of the Mc4r promoter by leptin were both attenuated, indicating that T3 or leptin may regulate Mc4r promoter activity by targeting the NKX2.1 sites on the Mc4r promoter (Figure 8).
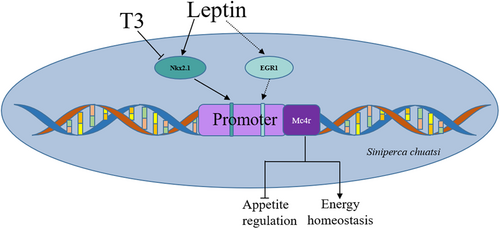
In addition, the activity of the −41 to +321 bp region of the Mc4r promoter was also significantly elevated after leptin incubation, which, in combination with previous TF predictions, revealed the presence of an early growth response factor-1 (EGR1) binding site in this region (Figure 2b). EGR1, also known as Zif/268, is typically activated by growth factors and cell depolarization (Sukhatme et al., 1988) and is also constitutively expressed in certain cell and neurons (Beckmann & Wilce, 1997). Leptin enhanced cholecystokinin (CCK) activity by promoting the synthesis of EGR1, while CCK reduced food intake in rats by stimulating gene expression of the satiety peptides cocaine and amphetamine (de Lartigue et al., 2010). Thus, following leptin incubation, the −41 to +321 bp activity of the Mc4r promoter is greatly increased, most likely because EGR1 mediates the transcriptional regulatory activity of leptin on Mc4r (Figure 8), but further studies are needed to confirm this.
In conclusion, the Mc4r promoter of mandarin fish was successfully cloned, and its structure and function were resolved. NKX2.1 was discovered to be involved in the transcriptional regulation of Mc4r and to mediate the stimulation of transcriptional activity of the Mc4r promoter by thyroid hormone and leptin for the first time in fish.
AUTHOR CONTRIBUTIONS
Di-Mei Xu: sample preparation, data analysis, complete experiment and wrote the manuscript. Jia-Qi Wu, Qiu-Ling Wang, and Xiao-Dan Jia: contributed to the sample preparation. Shan He and Xu-Fang Liang: gave technical advice and contributed to the study design. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This work was financially supported the National Natural Science Foundation of China (31972809) and the Key Research & Development Program of Hubei Province (2022BBA0051).