BMP-2 promotes differentiation of osteoblasts and chondroblasts in Runx2-deficient cell lines
Abstract
To investigate the molecular mechanism underlying the differentiation of osteoblasts and chondroblasts, we established a clonal cell lines, RD-C6, from Runx2-deficient mouse embryos. RD-C6 cells expressed almost undetectable levels of phenotypes related to osteoblast and chondroblast differentiation at basal culture condition, whereas treatment with recombinant human bone morphogenetic protein-2 (rhBMP-2) or transduction of BMP-2 by adenovirus effectively induced this cell line to express mRNA related to the differentiation of osteoblasts and chondroblasts including alkaline phosphatase, osteocalcin, and osterix. Transduction of Runx2 also induced the expression of these mRNA in RD-C6 cells. BMP-2 transduction increased expression levels of mRNA for Msx2 and Dlx5, but Runx2 transduction induced no significant increases in expression levels of these mRNA. Microarray analysis using RD-C6 cells with or without rhBMP-2 treatment demonstrated that BMP-2 upregulated 66 genes including 13 transcription-related molecules such as Id1, Id2, Id4, Hey1, Smad6, Smad7, and Msx2. To confirm bone and cartilage formation ability of RD-C6 cells, we transplanted RD-C6 cells into the peritoneal cavity of athymic mice using diffusion chambers with rhBMP-2. RD-C6 cells generated unmineralized cartilage but not bone. These results indicate that BMP-2 induces Runx2-deficient cells to express markers related to osteoblast and chondroblast differentiation using a Runx2-independent pathway, but it failed to induce these cells to differentiate into bone-forming osteoblasts and mature chondrocytes. J. Cell. Physiol. 211: 728–735, 2007. © 2007 Wiley-Liss, Inc.
Skeletal tissue is composed of various types of mesenchymal cells such as osteoblasts, chondrocytes, myocytes, and adipocytes. These cells originate from common pluripotent progenitors called mesenchymal stem cell (Caplan, 1991; Pittenger et al., 1999). During the differentiation of these cells, lineage-specific transcription factors critically regulate the pluripotent progenitors in their acquisition of specific phenotypes depending on their maturation (Rodan and Harada, 1997; Yamaguchi et al., 2000). Among these, Runx2 (runt-related gene 2, alternatively called core binding factor alpha 1: Cbfa1) is an essential transcription factor for osteoblast differentiation and bone formation (Ducy et al., 1997; Komori et al., 1997; Otto et al., 1997; Stein et al., 2004); this was understood based on the fact that Runx2-deficient mice completely lacked bone formation due to the maturation arrest of osteoblasts (Komori et al., 1997; Otto et al., 1997). These mice also displayed the delayed maturation of chondrocytes (Inada et al., 1999), indicating that Runx2 also regulates the differentiation and maturation of chondrocytes (Enomoto et al., 2000; Ueta et al., 2001; Yoshida et al., 2004). Osterix is another essential transcription factor that regulates osteoblast differentiation and bone formation (Nakashima et al., 2002). Osterix-deficient mice also exhibited lack of bone formation due to the maturation arrest of osteoblast differentiation, but they showed no apparent changes in cartilage (Nakashima et al., 2002). Thus, these two transcription factors govern the critical regulation of osteoblast differentiation and bone formation.
Bone morphogenetic proteins (BMPs) were originally identified as proteins that induced ectopic bone formation when implanted into muscular tissue (Urist, 1965), and they are strong inducers of osteoblast differentiation and bone formation (Yamaguchi et al., 1991, 2000; Katagiri et al., 1994). Several lines of evidence have demonstrated that BMP-2 induces or promotes the expression of mRNA for Runx2 (Ducy et al., 1997; Lee et al., 2000; Lee et al., 2003a) and osterix (Lee et al., 2003b; Celil and Campbell, 2005) as well as osteoblast differentiation markers such as alkaline phosphatase (ALP), type I collagen and osteocalcin in various cells. We have reported that the primary calvarial cells isolated from Runx2-deficient mouse embryos displayed extremely low or undetectable levels of osteoblast and chondroblast markers, whereas BMP-2 treatment induced these cells to express these markers (Komori et al., 1997; Kobayashi et al., 2000); this suggests that the differentiation of osteoblasts and chondroblasts is partly regulated by a Runx2-independent pathway. These results indicate that the cells isolated from Runx2-deficient mice are useful for developing an insight into the molecular events involved in a Runx2-independent pathway during osteoblast differentiation. There is, however, a limitation in using the primary cells isolated from Runx2-deficient mouse embryos for various experiments; this limitation is difficulty to isolate the cells for each experiment and maintain the characteristics of the cells for every experiment. This problem prompted us to establish new cell lines from the calvaria of Runx2-deficient mouse embryos in order to explore more distinct role of Runx2 in osteoblast and chondroblast differentiation.
In the present study, we established the clonal Runx2-deficient cell line, RD-C6, and investigated the potency of their differentiation into osteoblasts and chondroblasts. We demonstrate here that RD-C6 cells are capable of differentiating into certain stages of osteoblasts and chondroblasts by stimulation of BMP-2 or Runx2; this indicates the usefulness of this cell line for searching molecular events underlying osteoblast and chondroblast differentiation by Runx2-independent pathway.
MATERIALS AND METHODS
Isolation and culture of Runx2-deficient cell lines
The cells at calvarial region were isolated from Runx2-deficient mouse embryos as described previously (Komori et al., 1997). Briefly, fibrous connective tissues in the calvarial region of Runx2-deficient mice were dissected from day 18.5 embryos. They were washed in phosphate buffered saline (PBS) and cut into pieces, approximately 2 × 2 mm in size, and cultured for 14 days in three-dimensional collagen gel (Cell matrix, Nitta Gelation Co., Osaka, Japan) in α-modified minimum essential medium (α-MEM) containing 10% fetal bovine serum (FBS). The cells outgrowing from calvariae were retrieved by incubation for 30 min at 37°C with PBS containing 0.2% collagenase (Wako Pure Chemical Industries, Osaka, Japan) and 0.02% ethylenediaminetetraacetic acid (EDTA). The cells obtained were cultured with α-MEM containing 10% FBS (control medium), and then subcultured every 3 days until passage 40. Clonal cell lines were isolated from the 39th passage cells by a limiting dilution technique, and 10 clonal cell lines were established. Several cultures were treated with recombinant human BMP-2 (rhBMP-2), which was provided by Astellas Pharma, Inc., Tokyo, Japan, at various concentrations. One cell line, RD-C6, was chosen for further investigation based on the responsiveness to rhBMP-2. These cells and MC3T3-E1 cells were inoculated in culture dishes at a density of 2 × 104 cells /cm2 in every experiment. The genotypes of two cell lines were confirmed by Southern blot analysis using genomic DNA isolated from these cell lines as described previously (Komori et al., 1997).
Measurement of ALP activity
The cultured cells were sonicated in 0.1 M Tris buffer, PH7.2, containing 0.1% Triton X-100. ALP activity was determined using p-nitrophenylphosphate as a substrate in 50 mM 2-amino-2-methylpropanol and 2 mM MgCl2 with PH10.5 (Yamaguchi et al., 1991). The amount of p-nitrophenylphosphate released was estimated by measuring the absorbance at 410 nm. The protein concentration was determined using a BCA Protein Assay Kit (Pierce Chemical Co., Rockford, IL). ALP activity was also determined by a histochemical technique using naphtol AS-MX phosphate and fast blue BB salt (Katagiri et al., 1994).
Northern blot and reverse transcription-PCR analyses
The total RNA was extracted from the cultured cells for Northern blot analysis using RNA Smart Total RNA Isolation Kit (Nippon Genetics, Tokyo, Japan). Aliquots of 5 µg of total RNA were electrophoresed in 1% agarose formaldehyde gel and transferred onto a positively charged nylon transfer membrane (Amersham Biosciences, Buckinghamshire, UK). The membrane was UV-cross-linked, prehybridized in DIG Easy Hyb (Roche Diagnostics, Indianapolis, IN) for 30 min at 68°C, and then hybridized overnight at 68°C in Easy Hyb with DIG-labeled RNA probe for Runx2. After washing, the membrane was incubated for 30 min in blocking solution followed by 30 min incubation in anti-DIG-AP (Roche Diagnostics, Indianapolis, IN) diluted in blocking solution. After incubation, the membrane was washed twice in washing buffer, equilibrate in detection buffer, and the hybridized bands were detected by chemiluminescence using CDP-Star substrate (Roche Diagnostics, Indianapolis, IN). The blots were rehybridized with DIG-labeled GAPDH.
Aliquots of 400 ng RNA were reverse transcribed to cDNA using the 1st Strand cDNA Synthesis Kit for RT-PCR (Roche Diagnosis, Mannheim, Germany). The resulting cDNA products were subjected to PCR amplification using the gene-specific primers for ALP, osteocalcin, Col2a1, and β-actin. The number of cycles used in PCR was as follow: 36 cycles for ALP, 30 cycles for osteocalcin, 26 cycles for Col2a1, and 23 cycles for β-actin. The amplified products were electrophoresed in 2% agarose gel containing ethidium bromide with pUC Mix Marker, 8 (Fermentas, Hanover, MD). Alternatively, the expression of mRNA was quantified by real time-based RT-PCR using a Light Cycler System (Roche, Mannheim, Germany) with a Platinum SYBR Green qPCR SuperMix UDG Kit (Invitrogen, Carlsbad, CA). The relative amount of each mRNA was normalized by GAPDH expression in real time-based RT-PCR. The specific primers used are listed in Table 1.
| Forward primer | Reverse primer | |
|---|---|---|
| ALP | 5′-TGAGCGACACGGACAAGA | 5′-GGCCTGGTAGTTGTTGTGAG |
| Osteocalcin | 5′-CCAAGCAGGAGGGCAATA | 5′-AGGGCAGCACAGGTCCTAA |
| Col 2a1 | 5′-TGCACGAAACACACTGGTAAG | 5′-CACCAAATTCCTGTTCAGCC |
| β-actin | 5′- GCCACCAGTTCGCCATGGAT | 5′-AATGGGGTACTTCAGGGTCA |
| Osterix | 5′-TATGCTCCGACCTCCTCAACT | 5′-TCCTATTTGCCGTTTTCCCGA |
| Msx2 | 5′-AACACAAGACCAACCGGAAG | 5′-GCAGCCATTTTCAGCTTTTC |
| Dlx5 | 5′-CTGGCCGCTTTACAGAGAAG | 5′-CTGGTGACTGTGGCGAGTTA |
| Id4 | 5′-GTTCACGAGCATTCACCGTA | 5′-AAGGTTGGATTCACGATTGC |
| Hey1 | 5′-AGCAGTGAGGTGAAGGGAGA | 5′-AACGGTGAAATCCGTGAGAC |
| Grem2 | 5′-CCAGCCTCTGAGACTTCACC | 5′-ATTCAGCTGGTGGGTGTTTC |
| Latexin | 5′-GTCGCCTGCGGTTATGTAAT | 5′-GGCGGCTGTTGTGTTTTACT |
| GAPDH | 5′-TGACGGGAAGCTCACTGG | 5′-TCCACCACCCTGTTGCTGTA |
Transduction with adenovirus expressing BMP-2, Runx2, and GFP
RD-C6 cells were transduced with a replication-deficient adenoviral vectors carrying the human BMP-2 gene (AdBMP-2; Okubo et al., 1999) or the mouse Runx2 gene (AdRunx2; Yoshida et al., 2002). The recombinant adenovirus encoding green fluorescent protein (AdGFP) was used as control. The cells growing to 70% confluence in 6-well or 24-well plates were incubated with media containing an adenovirus (MOI: 400) according to the manufacturer's instructions (BD Biosciences Clontech, Alto, CA). The transfected cells were cultured for up to 6 days to analyze the differentiation of osteoblasts and chondroblasts.
Microarray analysis
The RNA sample for RNA microarray analysis was prepared from RD-C6 cells with or without rhBMP-2 treatment (500 ng/ml) for 2 days. cDNA was synthesized and then cRNA was labeled and amplified using a Low RNA Fluorescent Linear Amplification Kit (Agilent Technologies, Santa Clara, CA). The RNA from RD-C6 cells without rhBMP-2 treatment was labeled with Cy3, and that from RD-C6 cells treated with rhBMP-2 was labeled with Cy5. The labeled cRNA was purified by using RNeasy Mini Spin Columns (Qiagen, Valencia, CA). Hybridization was performed at 60°C for 17 h using a Whole Mouse Genome Oligo DNA Microarray (Agilent Technologies). These arrays contain probes for about 41,000 genes. The slides were scanned by an Agilent Technologies Microarray Scanner. Data were analyzed using Feature Extraction Software 8.1 (Agilent Technologies).
Transplantation of cells using diffusion chambers
Diffusion chambers (Millipore Corporation, Bedford, MA) were assembled as reported previously (Yamaguchi et al., 1996). A suspension of 2 × 106 RD-C6 cells in 100 µl of α-MEM containing 10% FBS was injected into each diffusion chamber coated with 2 µg rhBMP-2, cultured overnight, and then transplanted into the peritoneal cavity of athymic mice. Four weeks after the transplantation, the diffusion chamber was recovered and fixed in 10% buffered formalin in PBS. The dehydrated samples were embedded in Technovit 8100 (Heraeus Kulzer GmbH & Co., Wehrheim, Germany). Undecalcified sections of 4 µm were prepared and stained by toluidine blue, ALP or von Kossa's methods.
Statistics
Statistical analyses were performed using Student's unpaired t test. Each experiment was conducted at least twice, and representative data were presented as mean ± SD of independent replicates (n > 3).
RESULTS
Isolation and establishment of Runx-2-deficient cell lines
We isolated 10 clonal cell lines from the 39th passage of the maternal cell line, which was established from the calvarial region of Runx2-deficient mouse embryos. These clones showed various responses to rhBMP-2 treatment on ALP activity (Fig. 1A). Among these clones, we chose the clonal cell line, RD-C6, for further studies based on its high ALP activity induced by rhBMP-2 treatment. RD-C6 cells showed the highest response to rhBMP-2 by increasing ALP activity about 10-fold compared with non-treated RD-C6 cells (Fig. 1A). Through Southern blot analysis, we confirmed that the genotypes of Runx2 gene in 2 cell lines, RD-C2 and RD-C6, were the same as those in primary cells isolated from Runx2-deficient mice (Fig. 1B). Northern blot analysis also confirmed no expression of Runx2 mRNA in these cell lines with or without rhBMP-2 treatment (Fig. 1C).
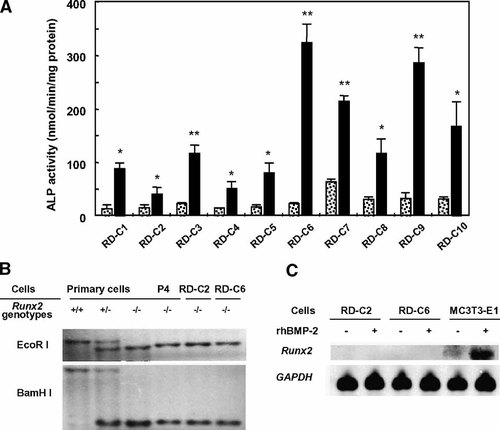
Isolation and establishment of Runx2-deficient cell lines. A: Effects of rhBMP-2 on ALP activity in 10 clonal cells isolated from the maternal Runx2-deficient cell line. Each cell line was treated with 500 ng/ml of rhBMP-2 for 6 days, and ALP activity was measured as described in Materials and Methods. Dotted bars represent ALP activity in the cells without rhBMP-2 treatment (control), and closed bars represent that in the cells treated with rhBMP-2. *P < 0.05 and **P < 0.01 versus control cells. B: Southern blot analysis. Primary cells: primary calvarial cells isolated from Runx2-deficient mouse embryos, P4: primary cells subcultured 4 times. +/+: Cells isolated from wild type mice, +/−: Cells isolated from heterozygous mice, −/−: cells isolated from Runx2-deficient mice. RD-C2 and RD-C6 are names of the clonal cell lines. C: Northern blot analysis for Runx2 mRNA. RD-C2, RD-C6, and MC3T3-E1 cells were treated with rhBMP-2 (500 ng/ml) for 3 days.
rhBMP-2 induced RD-C6 cells to express phenotypes related to osteoblast and chondroblast differentiation
We next investigated further the effects of rhBMP-2 on expression of several markers related to osteoblast and chondroblast differentiation using RD-C6 cells. RD-C6 cells cultured with control medium showed extremely low levels of ALP activity, whereas rhBMP-2 treatment for 6 days stimulated ALP activity in a dose-dependent fashion (Fig. 2A). RT-PCR analyses demonstrated that treatment with rhBMP-2 induced the expression of ALP mRNA on day 2 after the treatment with rhBMP-2 (Fig. 2B). RD-C6 cells expressed undetectable levels of osteocalcin mRNA at basal culture condition; however treatment with rhBMP-2 induced the apparent expression of osteocalcin mRNA on day 6. These expression profiles of mRNA for ALP and osteocalcin were quantitatively confirmed by real time-based RT-PCR analysis (data not shown).
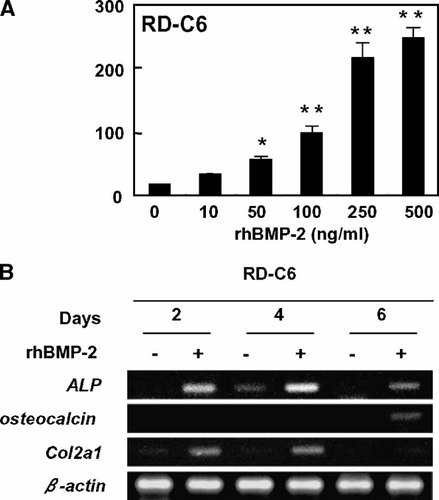
Treatment with rhBMP-2 induces RD-C6 cells to express markers related to osteoblast and chondroblast differentiation. A: Dose response effects of rhBMP-2 on ALP activity in RD-C6 cells. These cells were treated with the indicated doses of rhBMP-2 for 6 days, and ALP activity was measured as described in Materials and Methods. *P < 0.05 and **P < 0.01 versus control cells. B: RT-PCR analyses for mRNA expression of ALP, osteocalcin, and Col2a1. RD-C6 cells were treated with 500 ng/ml of rhBMP-2 for 2, 4, and 6 days. RT-PCR analysis was conducted as described in Materials and Methods.
RD-C6 cells expressed extremely low levels of mRNA for Col2a1, which is a marker of immature chondroblasts, in RD-C6 cells at basal culture condition. Treatment with rhBMP-2 increased the expression level of Col2a1 mRNA as early as on day 2 in RD-C6 cells (Fig. 2B), and it diminished on day 6. Treatment with rhBMP-2 failed to induce the apparent expression of mRNA for Col10a1, a marker of mature chondrocytes, in RD-C6 cells by RT-PCR analysis (data not shown).
Overexpression of Runx2 and BMP-2 induced osteoblast and chondroblast differentiation in RD-C6 cells
To investigate whether exogenous overexpression of Runx2 could rescue the differentiation of osteoblasts and chondroblasts in RD-C6 cells, we transduced Runx2 using adenovirus, and compared the effects with those of transduction with BMP-2 or GFP. Transduction with BMP-2 induced a greater number of ALP-positive cells than that with Runx2 transduction in RD-C6 cells (Fig. 3A). Transduction with Runx2 induced the expression of osteocalcin mRNA more effectively than that with BMP-2 in RD-C6 cells (Fig. 3B). Transduction with BMP-2 induced osterix more effectively than that with Runx2 during the culture period (Fig. 3B). Transduction with BMP-2 significantly increased the expression levels of Msx2 mRNA during the culture period in RD-C6 cells (Fig. 3B). Transduction with Runx2, however, induced no significant increase in the expression level of Msx2 mRNA. BMP-2 transduction upregulated Dlx5 expression on days 2 and 4, but Runx2 transduction induced no significant changes in the expression level of Dlx5 mRNA (Fig. 3B).
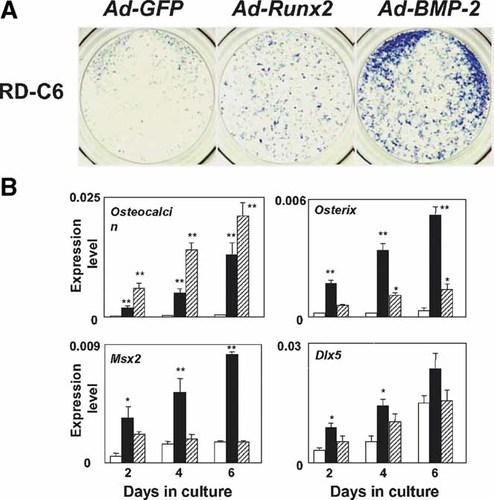
Effects of transduction with Runx2 and BMP-2 by adenovirus on osteoblast phenotypes in RD-C6 cells. A: RD-C6 cells were transduced with Runx2 and BMP-2 as described in Materials and Methods. ALP activity was detected histochemically as described in Materials and Methods on day 6 after transduction with each gene. B: Effects of transduction with Runx2 and BMP-2 by adenovirus on expression of mRNA for osteocalcin, osterix, Msx2, and Dlx5. The expression of mRNA was assessed by real time-based RT-PCR analysis using RD-C6 cells on days 2, 4, and 6 after transduction with Runx2 and BMP-2. Open bars represent the expression levels of mRNA in RD-C6 cells transduced with Ad-GFP (control cells). Closed bars show levels of mRNA in RD-C6 cells transduced with Ad-BMP-2, and shadow bars represent that in the cells transduced with Ad-Runx2; *P < 0.05 and **P < 0.01 versus control cells. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Transduction with Runx2 and BMP-2 increased the expression of Col2a1 mRNA during culture period in RD-C6 cell (data not shown). Transduction with Runx2 and BMP-2 slightly increased the expression level of Col10a1 mRNA on days 4 and 6, although its expression level was extremely low (data not shown).
Expression profiles of BMP-2-induced genes in RD-C6 cells by microarray analysis
Since rhBMP-2 induced expression of mRNA related to osteoblast differentiation in RD-C6 cells, we conducted microarray analysis using these cells with or without rhBMP-2 treatment for 2 days to characterize the expression profiles of the genes induced by rhBMP-2 treatment.
rhBMP-2 treatment upregulated 66 genes over 2-fold as shown in Table 2. These genes were divided into subgroups according to the function. We detected 13 genes related to transcription. Among them, Id4 was the most highly upregulated (14.6-fold; Table 2). As shown in Figure 4, the expression level of Id4 greatly increased by rhBMP-2 treatment on day 2 as well as days 4 and 6 in RD-C6 cells by real time-based RT-PCR analysis. The other Id family genes, Id1 and Id2, were also upregulated by 3.08-fold and 2.92-fold, respectively (Table 2). Real time-based RT-PCR analyses revealed that BMP-2 stimulation significantly increased the expression levels of mRNA for Id2 and Id3 in RD-C6 cells on days 2 and 4 in culture (data not shown).
| Gene name | Description | Gene bank accession no. | Change fold |
|---|---|---|---|
| Transcription factors (13) | |||
| Id4 | Inhibitor of DNA binding 4 | NM_031166 | 14.6 |
| Hey1 | Hairy/enhancer-of-split related with YRPW motif 1 | NM_010423 | 13.8 |
| Smad7 | MAD homolog 7 (Drosophila) | NM_008543 | 6.59 |
| Foxo6 | Forkhead box O6 | NM_194060 | 5.72 |
| Smad6 | MAD homolog 6 (Drosophila) | NM_008542 | 4.03 |
| Foxf2 | Forkhead box F2 | NM_010225 | 3.28 |
| Id1 | Inhibitor of DNA binding 1 | NM_010495 | 3.08 |
| Id2 | Inhibitor of DNA binding 2 | NM_010496 | 2.92 |
| Irf5 | Interferon regulatory factor 5 | NM_012057 | 2.87 |
| Msx2 | Homeobox, msh-like 2 | NM_013601 | 2.73 |
| Snai1 | Snai homolog 1 | NM_001427 | 2.45 |
| Maff | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F | NM_010755 | 2.38 |
| Dlx2 | Distal-less homeobox 2 | NM_010054 | 2.38 |
| Growth factors (8) | |||
| Grem2 | Gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) | NM_011825 | 14.5 |
| Igfbp5 | Insulin-like growth factor binding protein 5 | BC003951 | 5.22 |
| Fst | Follistatin | NM_008046 | 4.18 |
| Mrpplf4 | Mitogen regulated protein, proliferin 4 | NM_181852 | 3.54 |
| Plf2 | Proliferin2 | NM_011118 | 3.38 |
| Ctgf | Connective tissue growth factor | NM_010217 | 2.80 |
| Cyr61 | Cysteine rich protein 61 | NM_010516 | 2.26 |
| Inhba | Inhibin beta-A | NM_008380 | 2.14 |
| Receptors (7) | |||
| Unc5b | Unc-5 homolog B | NM_029770 | 8.26 |
| Cxcr6 | Chemokine (C-X-C motif) receptor 6 | NM_030712 | 3.58 |
| Gpr48 | G protein-coupled receptor 48 | AK044357 | 3.45 |
| Tmeff2 | Transmembrane protein with EGF-like and two follistatin-like domains 2 | NM_019790 | 3.23 |
| Temff1 | Transmembrane protein with EGF-like and two follistatin-like domains 1 | NM_021436 | 2.66 |
| Trem2 | Triggering receptor expressed on myeloid cells 2 | NM_031254 | 2.71 |
| Ahr | Aryl-hydrocarbon receptor | NM_013464 | 2.14 |
| Signal transduction (3) | |||
| Wnt2 | Wingless-related MMTV integration site 2 | NM_023653 | 3.29 |
| Mcf2l | Mcf.2 transforming sequence-like | NM_178076 | 2.40 |
| Wisp2 | WNT1 inducible signaling pathway protein 2 | NM_016873 | 2.22 |
| Extracellular matrix/cell adhesion molecules (7) | |||
| Smoc1 | SPARC related modular calcium binding 1 | NM_022316 | 4.22 |
| Cthrc1 | Hypothetical collagen triple helix repeat containing protein | AK003674 | 4.08 |
| Itgb6 | Integrin beta 6, cell adhesion and cell-matrix adhesion | NM_021359 | 3.88 |
| Masp1 | Mannan-binding lectin serine protease | AK031598 | 3.45 |
| Lum | Lumican | NM_008524 | 3.07 |
| Cdsn | Corneodesmosin | NM_001008424 | 2.72 |
| Fmod | Fibromodulin | NM_021355 | 2.65 |
| Structural/cytoskeletal proteins (4) | |||
| Lor | Loricrin | NM_008508 | 4.23 |
| Cnn1 | Calponin | NM_009922 | 2.96 |
| Golph4 | Golgi phosphoprotein 4 | NM_175193 | 2.61 |
| Pkp1 | Plakophilin 1 | NM_019645 | 2.38 |
| Others (24) | |||
| Akp2 | Alkaline phosphatase 2 | NM_007431 | 23.5 |
| Lxn | Latexin (Lxn) | NM_016753 | 8.93 |
| Has2 | Hyaluronan synthase 2 | NM_008216 | 5.57 |
| Greb1 | Gene regulated by estrogen in breast cancer protein | NM_015764 | 4.59 |
| Pappa | Pregnancy-associated plasma protein A, | NM_021362 | 3.76 |
| Serpine1 | Serine (or cysteine) proteinase inhibitor, clade E, member 1 (Serpine1) | NM_008871 | 3.76 |
| Sytl2 | Synaptotagmin-like 2, membrane fraction | NM_031394 | 3.27 |
| Aldh 1a7 | Aldehyde dehydrogenase family 1, subfamily A7 | NM_011921 | 3.26 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | NM_011198 | 3.22 |
| Fabp7 | Fatty acid binding protein 7 | NM_021272 | 3.19 |
| Gse1 | Genetic suppressor element 1 | NM_198671 | 3.02 |
| Baalc | Brain and acute leukemia, cytoplamic, function unknown | NM_080640 | 3.02 |
| Gadd45b | Growth arrest and DNA-damage-inducible 45 beta | NM_008655 | 2.95 |
| Pcolce2 | Procollagen C-endopeptidase enhancer 2 | NM_029620 | 2.84 |
| Cyp26b1 | Cytochrome P450, family 26, subfamily b, polypeptide 1 | NM_175475 | 2.83 |
| Aass | Aminoadipate-semialdehyde synthase | NM_013930 | 2.69 |
| Rif1 | Rap1 interacting factor 1 homolog (yeast) | NM_175238 | 2.65 |
| Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 | NM_201531 | 2.61 |
| Usmg4 | Upregulated during skeletal muscle growth 4 | NM_031401 | 2.55 |
| Adamts9 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 2, motif 9 | BC068142 | 2.55 |
| Bpgm | 2,3-bisphosphoglycerate mutase | NM_007563 | 2.52 |
| Psph | Phosphoserine phosphatase | NM_133900 | 2.36 |
| Nrn1 | Neuritin 1 | NM_153529 | 2.33 |
| Enah | Enabled homolog | NM_010135 | 2.33 |
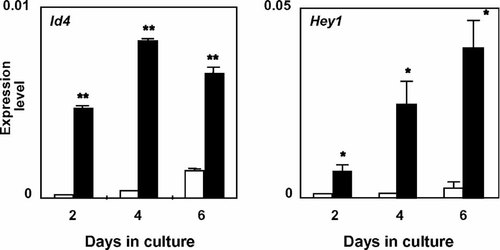
Effects of rhBMP-2 on the expression of mRNA for Id4 and Hey1 in RD-C6. The cells were treated with rhBMP-2 (500 ng/ml) for 2, 4, and 6 days. The expression of each mRNA was assessed by real time-based RT-PCR analysis. Open bars represent the expression levels of each mRNA in the cells without rhBMP-2 treatment (control), and closed bars represent that in the cells treated with rhBMP-2; *P < 0.05 and **P < 0.01 versus control cells.
Hey1, a transcription factor belonging to the basic helix-loop-helix superfamily, was also highly upregulated by 13.8-fold (Table 2). As shown in Figure 4, real time-based RT-PCR analysis confirmed that treatment with rhBMP-2 significantly stimulated the expression level of Hey1 during the culture period in RD-C6 cells. Two homeobox genes, Msx2 and Dlx2, were also upregulated by 2.73-fold and 2.38-fold, respectively. Two inhibitory Smads, Smad 6, and Smad 7, showed upregulation with 4.03-fold and 6.59-fold, respectively. Upregulation of these two mRNA during culture period in RD-C6 cells was also confirmed by real time-based RT-PCR (data not shown). Two genes belonging to the forkhead/winged helix loop family, Foxo6 and Foxf2, were increased over 3-fold.
The genes belonging to the growth factor-related group consisted of seven genes. Grem2, which is a BMP antagonist, showed the highest level in this group with a 14.5-fold increase. Another BMP antagonist, follistatin (Fst) also exhibited upregulation by 4.18-fold. Two CCN family members, Ctgf and Cyr61, were both upregulated over 2-fold. Other growth factor-related genes include Igfbp5, Mrpplf4, and Plf2. Upregulated genes in the receptor group include Unc5b, Cxcr6, Gpr48, Tmeff1, Tmeff2, and Trem2. Three genes, Wnt2, Mcf21, and Wisp2, related to signal transduction were also upregulated over 2-fold. Seven genes, Smoc1, Cthrc1, Itgb6, Masp1, Lum, Cdsn, and Fmod, involved in extra cellular matrices and cell adhesion were upregulated over 2-fold. Four genes, Lor, Cnn1, Golph4, and Pkp1, associated with structural and cytoskeletal proteins exhibited over 2-fold increase. Among the 24 genes belonging to the other group, ALP showed the highest upregulation by 23.5-fold, and latexin exhibited a 8.93-fold increase.
RD-C6 cells form cartilage but not bone in response to BMP-2 in diffusion chambers
RD-C6 cells transplanted without rhBMP-2 for 4 weeks generated neither cartilage nor bone in the diffusion chambers (Fig. 5A). In contrast, RD-C6 cells transplanted with rhBMP-2 for 4 weeks formed numerous cartilage foci in the diffusion chambers (Fig. 5B). The cells located in this area showed weak ALP activity, but no mineralization was confirmed at cartilage foci by von Kossa stain (Fig. 5B). Some fibroblast-like cells adjacent to the filter membrane exhibited weak ALP activity (data not shown). These cells resembled osteoblastic cells, but they produced no mineralized bone matrix. Thus, RD-C6 cells are capable of generating cartilage in response to BMP-2, but they fail to generate bone in the diffusion chambers.
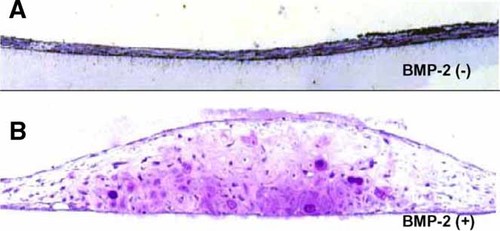
Histology of RD-C6 cells after transplantation into the peritoneal cavity of athymic mice with (B) or without (A) rhBMP-2 treatment as described in Materials and Methods. The sections were stained with toliudine blue following von Kossa staining. Note that no bones and mineralized cartilage were formed. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
We demonstrated by establishment of the Runx2-deficient cell line, RD-C6 cells, that BMP-2 is capable of inducing osteoblast differentiation by a Runx2-independent pathway. BMP-2 also enhanced the expression of Col2a1 mRNA in RD-C6 cells. Further, experiments using diffusion chambers demonstrated that RD-C6 cells are capable of generating cartilage in response to rhBMP-2. These results demonstrate that RD-C6 cells are capable of differentiating into certain stages of osteoblasts and chondroblasts in response to BMP-2.
Osterix is a candidate gene involved in osteoblast differentiation in Runx2-deficient cells. We found that the expression of osterix mRNA is upregulated by overexpression of BMP-2 and Runx2 in RD-C6 cells, suggesting that the expression of osterix mRNA is regulated by both Runx2-dependent and Runx2-independent pathways. Since the transduction of osterix induced the expression of ALP mRNA in RD-C6 cells as well as in C3H10T1/2 cells (unpublished results by Dr. Riko Nishimura), osterix might be one of the genes participating in the Runx2-independent pathway. The other candidate genes participating in this pathway are the two homeobox genes, Msx2 and Dlx5; this inference was drown from several reports that demonstrated that these genes are involved in osteoblast differentiation (Ryoo et al., 1997; Miyama et al., 1999; Cheng et al., 2003; Lee et al., 2003a,b; Ichida et al., 2004; Yoshizawa et al., 2004; Ryoo et al., 2006). In the present study, we have revealed that BMP-2 effectively increased the expression level of Msx2 mRNA in RD-C6 cells, but Runx2 transduction induces no significant changes in its expression. These results indicate that Msx2 is a downstream factor of BMP-2 signaling, but not Runx2. Further, we observed that the transduction of Msx2 promotes osteoblast differentiation in not only C3H10T1/2 cells but also Runx2-deficient primary cells (Ichida et al., 2004). These results indicate that Msx2 participates in transcriptional machinery in the Runx2-independent pathway during osteoblast differentiation. Lee et al. (2003b) suggested that osterix is a downstream target of Dlx5 during BMP-2-induced osteoblast differentiation. The present study revealed that BMP-2 promotes the expression level of Dlx5, but Runx2 does not, in RD-C6 cells. In contrast, Dlx5 expression is upregulated by Runx2 transduction, but not by BMP-2 transduction in RD-C2 cells, which is another Runx2-deficient cell line isolated from Runx2-deficcient mice (Fig. 1; unpublished results). These results suggest that both BMP-2 and Runx2 can regulate the expression of Dlx5. Thus, the present study suggests that osterix, Msx2, and Dlx5 participate in the Runx2-independent pathway in BMP-2-induced osteoblast differentiation.
We confirmed 66 genes upregulated over 2-fold by rhBMP-2 treatment by microarray analysis using RD-C6 cells treated with or without rhBMP-2. Almost these genes identified were reported to be upregulated by BMP treatment in other osteogenic cells by microarray analyses, indicating that BMP can regulate the expression of these genes through Runx2-independent pathway. Among these, however, expression of Id4 mRNA displayed an opposite result to the previous report (Zamurovic et al., 2004). We noticed by microarray analysis that rhBMP-2 treatment upregulated the expression of Id4 mRNA in RD-C6 cells, although Zamurovic et al. (2004) reported that BMP-2 treatment inhibited its expression in MC3T3-E1 cells. We also confirmed by real time-based RT-PCR analyses that rhBMP-2 treatment upregulated the expression of Id4 mRNA in RD-C6 cells (Fig. 4), and downregulated its expression in MC3T3-E1 cells (unpublished results). These results indicate that BMP-2 has different effects on the expression of Id4 mRNA depending on the osteoblastic cells used. These results also suggest that Runx2 exerts an inhibitory effect on Id4 expression after BMP stimulation. Since the role of Id4 in osteoblast differentiation have not been clarified, it will be important to investigate the regulatory mechanism of Id4 during BMP-induced osteoblast differentiation.
Upregulation of Hey1 mRNA by BMP-2 treatment had been reported by microarray analyses using MC3T3-E1 (Zamurovic et al., 2004) and C2C12 (de Jong et al., 2004a,b) cells. We also demonstrated that the expression of Hey1 mRNA is upregulated by BMP-2 stimulation in Runx2-deficient cells (Table 2 and Fig. 4). These indicate that the expression of Hey1 mRNA is regulated through the Runx2-independent pathway during BMP-induced osteoblast differentiation. Hey1 is a negative regulator of osteoblast differentiation because it decreased Runx2 transcriptional activity (Zamurovic et al., 2004). Since Hey1 is a direct Notch target gene and we reported that Notch signaling plays critical roles in BMP-2-induced osteoblast differentiation (Nobta et al., 2005), further studies on the role of Hey1 during osteoblast differentiation are required.
The present study demonstrated that BMP stimulation promotes the expression of Col2a1 mRNA in RD-C6 cells, but failed to induce a substantial level of Col10a1 mRNA. These results suggest that Runx2-deficient cells retain the potential to differentiate into chondrocytes in response to BMP-2, although their ability to differentiate into mature chondrocytes might be very low. Since Runx2 is an important positive regulatory factor in chondrocyte maturation (Enomoto et al., 2000; Ueta et al., 2001; Yoshida et al., 2004), we transduced Runx2 into RD-C6 cells; however, it failed to induce an apparent expression of Col10a1 mRNA. This might be due to the lack of other essential transcription factors necessary for chondrocyte differentiation. We demonstrated that RD-C6 cells formed cartilage in response to rhBMP-2 in diffusion chambers when transplanted into the peritoneal cavity of athymic mice; however, the cartilage lacked mineralization, which is a marker of the terminal differentiation of chondrocytes. In short, BMP-2 can induce chondroblast differentiation in the absence of Runx2, but Runx2 plays a critical role in the terminal differentiation of chondrocytes (Enomoto et al., 2000; Ueta et al., 2001; Yoshida et al., 2004).
In summary, we demonstrated that BMP-2 induces Runx2-deficient cells to express markers related to osteoblast and chondroblast differentiation using the Runx2-independent pathway, but it fails to induce these cells to differentiate into bone-forming osteoblasts and mature chondrocytes. This indicates that Runx2 plays an essential role in osteogenesis and exerts a functional role in cartilage maturation. The Runx2-deficient cell line shown in this study will be a useful tool for searching the molecular events involved in Runx2-independent pathway during osteoblast and chondroblast differentiation, though it will be an important issue to consider whether the lack of the terminal differentiation of these cell lines are due to Runx2 deficiency and/or chromosomal abnormality of appropriate gene expression during establishment of the clonal cell lines.
Acknowledgements
We thank Dr. Riko Nishimura for providing information about unpublished results. This study was supported by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (14104015 to A.Y.).




