Transactivation of Src, PDGF receptor, and Akt is involved in IL-1β-induced ICAM-1 expression in A549 cells
Abstract
In previous study, interleukin-1β (IL-1β) has been shown to induce ICAM-1 expression through MAPKs and NF-κB in A549 cells. In addition to these pathways, transactivation of non-receptor tyrosine kinase (Src), PDGF receptors (PDGFRs), and phosphatidylinositol 3-kinase (PI3K)/Akt has been implicated in the expression of inflammatory genes. Here, we further investigated whether these different mechanisms participating in IL-1β-induced ICAM-1 expression in A549 cells. We initially observed that IL-1β-induced ICAM-1 promoter activity was attenuated by the inhibitors of Src (PP1), PDGFR (AG1296), PI3-K (LY294002 and wortmannin), and Akt (SH-5), revealed by reporter gene assay, Western blotting, and RT-PCR analyses. The involvement of Src and PI3-K/Akt in IL-1β-induced ICAM-1 expression was significantly attenuated by transfection of A549 cells with dominant negative plasmids of Src, p85 and Akt, respectively. Src, PDGFR, and PI3K/Akt mediated the effects of IL-1β because pretreatment with PP1, AG1296, and wortmannin also abrogated IL-1β-stimulated Src, PDGFR, and Akt phosphorylation, respectively. Moreover, pretreatment with p300 inhibitor (curcumin) also blocked ICAM-1 expression. We further confirmed that p300 was associated with ICAM-1 promoter which was dynamically linked to histone H4 acetylation stimulated by IL-1β, determined by chromatin immunoprecipitation assay. Association of p300 and histone-H4 to ICAM-1 promoter was inhibited by LY294002. Up-regulation of ICAM-1 enhanced the adhesion of neutrophils onto A549 cell monolayer exposed to IL-1β, which was inhibited by PP1, AG1296, LY294002, wortmannin, and helenalin. These results suggested that Akt phosphorylation mediated through transactivation of Src/PDGFR promotes the transcriptional p300 activity and eventually leads to ICAM-1 expression induced by IL-1β. J. Cell. Physiol. 211: 771–780, 2007. © 2007 Wiley-Liss, Inc.
Lung inflammation is a pivotal event in the pathogenesis of chronic obstructive pulmonary disease (Szulakowski et al., 2006), asthma (Barnes et al., 2005), and severe acute respiratory syndrome (SARS) (Dandekar and Perlman, 2005). These inflammatory responses are mediated by complex interactions between both circulating polymorphonuclear cells (PMNs) and the vascular endothelium. Several studies indicate that expression of adhesive molecules on the cell surface of endothelial cells plays a critical role in the inflammatory responses (Walzog and Gaehtgens, 2000). For example, patients with lung diseases have an increased adhesion activity linked to adhesive molecule between the epithelial cells and PMNs (Lazaar and Panettieri, 2005; Yang et al., 2005; Armstrong et al., 2006). Raised levels of adhesive molecules might contribute to the recruitment of PMNs to the regions of inflammatory tissue. These adhesion molecules are classified into two major families: the Ig superfamily (e.g., ICAM-1 and VCAM-1) and the selectins (e.g., P-selectin and E-selectin) (Min et al., 2005). ICAM-1 is an inducible cell surface glycoprotein on several cell types, which mediates the tight adhesiveness of PMNs and thus facilitates PMNs migration across the vascular endothelium barrier and then interacts with lung epithelium (Lin et al., 2005). Up-regulation of ICAM-1 on the surface of endothelial cells or multiple airway resident cells is prominent when they are exposed to proinflammatory molecules such as TNF-α, LPS, and vascular endothelial growth factor (Kim et al., 2001; Javaid et al., 2003; Min et al., 2005; Yang et al., 2005; Basit et al., 2006). Thus, to clarify mechanisms of ICAM-1 induction by interleukin-1β (IL-1β) in lung epithelium was recognized as a new therapeutic approach in the management of respiratory diseases.
Up-regulation of ICAM-1 by IL-1β has been shown to be mediated through activation of MAPKs and NF-κB in A549 cells (Lin et al., 2005). In addition to these pathways, the non-receptor tyrosine kinases (Src), PDGF receptor (PDGFR), and phosphatidylinositol 3-kinase (PI3-K)/Akt have been implicated in linking a variety of G-protein coupled receptors to expression of inflammatory genes. However, little was known about the way whether IL-1R shared the same mechanisms mediated through Src and PDGFR as intermediate components thereby mimicking the signaling pathways downstream of PDGFR. A previous study has shown that activation of IL-1R leads to phosphorylation of several substrates on tyrosine residue such as Src (al-Ramadi et al., 2001). c-Src is a multidomain protein and consists of an N-terminal membrane localization SH4 region, an SH3 domain, an SH2 domain linked to the kinase domain, and a C-terminal tail (Schlessinger, 2000; Bromann et al., 2004). In quiescent cells, a number of intramolecular forces maintain c-Src in its inactive state. A C-terminal phosphotyrosine binds to the SH2 domain, resulting in its unavailability to associate with other tyrosine phosphorylated signaling proteins. Although Src activity is thought to be necessary for transactivation of PDGFR by GPCRs (Tanimoto et al., 2004) and may lead to stimulate two major kinase cascades: Ras/Raf/MEK/ERK1/2 and PI3-K/Akt (Heldin and Westermark, 1999). Enhanced activity of PI3K/Akt by cytokines may induce expression of pro-inflammatory mediators and then contribute to the pathogenesis of asthma in the recruitment and activation of inflammatory cells (Datta et al., 1999; Kwak et al., 2003). In fact, phosphorylation of Akt may eventually regulate NF-κB translocation (Datta et al., 1999) and stimulate histone acetyltransferases (HAT) activity (Darieva et al., 2004). The promoter region of human ICAM-1 has been cloned and sequenced, and shown to contain putative recognition sequences for various transcriptional factors, including NF-κB and AP-1 (Voraberger et al., 1991). The NF-κB family proteins appear to be essential components for the enhanced ICAM-1 gene expression upon exposure to cytokines (Lin et al., 2005; Min et al., 2005). In addition, NF-κB-dependent ICAM-1 gene transcription is required the presence of HAT co-activators in cytokine-stimulated cells (Huang and Chen, 2005). HATs, such as p300 and CBP, which are two transcriptional co-activators, have been demonstrated to play central roles in coordinating and integrating multiple signal-dependent events with the transcriptional apparatus, allowing the appropriate level of gene activity to occur in response to diverse external stimuli. Previous studies have documented that Akt phosphorylation induces recruitment of p300 to the inflammation gene promoters, including COX-2, MMP-9, VCAM-1, and ICAM-1 (Ma et al., 2004; Takada et al., 2004; Huang and Chen, 2005; Lee et al., 2006) and leads to acetylation of histones in chromatin and association with the basal transcriptional machinery RNA polymerase II.
In the present study, we investigated that Akt was phosphorylated by Src and PDGFR transactivation stimulated by IL-1β. Akt phosphorylation further regulated p300 activity, recruited the transcriptional machinery to the ICAM-1 promoter and finally enhanced ICAM-1 expression in A549 cells.
Materials and Methods
Materials
DMEM/F-12 medium, FBS, TRIZOL, and Plus-Lipofectamine were purchased from Invitrogen (Carlsbad, CA). Hybond C membrane, enhanced chemiluminescence (ECL) Western blotting detection system, and Hyperfilms were from Amersham Biosciences (Buckinghamshire, England). Anti-ICAM-1, anti-p300, anti-acetyl H3, and anti-NF-κB p65 antibodies were from Santa Cruz (Santa Cruz, CA). PhosphoPlus Src, PhosphoPlus PDGFR, and PhosphoPlus Akt Ab kits were from Cell Signaling (Beverly, MA). Anti-GAPDH antibody was from Biogenesis (Boumemouth, UK). LY294002, wortmannin, curcumin, PP1, AG1478, AG1296, U0126, SB202190, SP600125, GM6001, and helenalin were from Biomol (Plymouth Meeting, PA). SH-5 was from Calbiochem (La Jolla, CA). Bicinchoninic acid (BCA) protein assay kit was from Pierce (Rockford, IL). Enzymes and other chemicals were from Sigma (St. Louis, MO).
Cell culture of A549
A549 cells, a human alveolar epithelial cell carcinoma, were purchased from the American Type Culture Collection (Manassas, VA) and cultured in DMEM/F-12 supplemented with 10% FBS and antibiotics (100 U/ml penicillin G, 100 µg/ml streptomycin, and 250 ng/ml fungizone) at 37°C in a humidified 5% CO2 atmosphere. When the cultures reach confluence (5 days), cells were treated with 0.05% (w/v) trypsin/1 mM EDTA for 5 min at 37°C. The cell suspension diluted with DMEM/F-12 containing 10% FBS to a concentration of 2 × 105 cells/ml. The cell suspension was plated onto (1 ml/well) 12-well culture plates and (10 ml/dish) 10-cm culture dishes for the measurement of protein expression and mRNA accumulation, respectively. Culture medium was changed after 24 h and then every 3 days.
Plasmids and transfection
The plasmids encoding dominant negative mutants of MEK1/2 (MEK K97R), ERK2 (ERK K52R), JNK, p38, Src, Akt, and p85 were kindly provided by Drs. K. L. Guan (Department of Biological Chemistry, University of Michigan), M. H. Cobb (Department of Pharmacology, University of Texas Southwestern Medical Center), J. Han (The Scripps Research Institute, La Jolla, CA, USA), C. C. Chen (Department of Pharmacology, National Taiwan University, Taipei, Taiwan), and R. D. Ye (Department of Pharmacology, University of Illinois at Chicago, USA), respectively. All plasmids were prepared by using QIAGEN plasmid DNA preparation kits.
A549 cells were plated at 3 × 105 cells/ml (1 ml/well) in 12-well culture plates for 24 h, reaching about 80% confluence. Cells were washed once with PBS and replaced with 0.4 ml of serum free DMEM/F-12 medium to each well. The DNA Plus-Lipofectamine reagent complex was prepared according to the instructions of manufacturer. The amount of plasmid transfected was kept constant (1 µg of dominant negative mutant for each well). The DNA PLUS-Lipofectamine reagent complex (0.1 ml) was added to each well and incubated at 37°C for 3 h, and then 1 ml of DMEM/F-12 medium containing 10% FBS was added and incubated for 21 h. After 24 h of transfection, the cells were washed twice with PBS and maintained in serum-free DMEM/F-12 for 24 h before treatment with IL-1β.
Preparation of cell extracts and Western blotting analysis
A549 cells were plated onto 12-well culture plates and made quiescent at confluence by incubation in serum-free DMEM/F-12 for 24 h. Growth-arrested cells were incubated with different concentrations of IL-1β at 37°C for the indicated time. When inhibitors were used, they were added 1 h prior to application of IL-1β. The highest concentrations of these inhibitors used did not cause any toxic effect on A549 cells (data not shown). After incubation, the cells were then rapidly washed with ice-cold PBS, scraped, and collected by centrifugation at 1,000g for 10 min. The collected cells were prepared as previously described (Lin et al., 2005). Samples from these supernatant fractions (30 µg protein) were denatured and subjected to SDS–PAGE using a 10% running gel. Proteins were transferred to nitrocellulose membrane and the membranes were incubated overnight at 4°C with anti-ICAM-1, anti-phospho-Src, anti-phospho-PDGFR, anti-phospho-Akt or anti-GAPDH Ab used at a dilution of 1:2,000 in TTBS. Membranes were washed with TTBS four times for 15 min each, incubated with a 1:1,500 dilution of anti-goat or anti-mouse horseradish peroxidase Ab for 1 h. Following each incubation, the membrane was washed extensively with TTBS. The immunoreactive bands detected by ECL reagents were developed by Hyperfilm-ECL.
Total RNA extraction and RT-PCR analysis
Total RNA was isolated from A549 cells exposed to IL-1β for the indicated time in 10-cm culture dishes with TRIZOL according to the protocol of the manufacturer. RNA concentration was spectrophometrically determined at 260 nm. The extents of ICAM-1 and β-actin RNA expression were determined by RT-PCR as previously described (Lin et al., 2005).
Luciferase activity assay
A549 cells, grown to 50% confluency in 6-well plates, were transfected with the human ICAM-1(pIC-339/0)/firefly luciferase (Luc) which was kindly provided by Dr. P. T. van der Saag (Hubrecht Laboratory, Utrecht, The Netherlands) or κB-luc plasmid using Metafectene transfection reagent was from Biontex (Munich, Germany) according to the manufacturer's instructions. Briefly, reporter DNA (0.4 µg) and β-galactosidase DNA (0.2 µg) were mixed with 0.6 µl of Tfx-50 in 1 ml of serum-free DMEM/F-12. After 15-min incubation at room temperature, the mixture was applied to the cells, and then 1 h later, 1 ml of complete growth medium was added. Transfection for 24 h, the cells were shifted to serum-free DMEM/F-12 medium for 24 h, treated with inhibitors (as indicated) for 60 min, and then IL-1β was added for 5 h. Cell extracts were then prepared and luciferase and β-galactosidase activities were measured. Firefly luciferase activities were standardized for β-galactosidase activity.
Chromatin immunoprecipitation assay
To detect the in vivo association of nuclear proteins with human ICAM-1 promoter, chromatin immunoprecipitation (ChIP) analysis was conducted as previously described (Nie et al., 2003) with some modifications. DNA immunoprecipitated by anti-p300 (sc-585; Santa Cruz), anti-acetylated histone H3 (06–599; Upstate Biotechnology, Lake Placid, NY), or anti-NF-κB p65 (sc-372; Santa Cruz) antibody was purified. The DNA pellet was re-suspended in H2O and subjected to PCR amplification with the forward primer 5′-AGACCTTAGCGCGGTGTAGA-3′ and the reverse primer 5′-AGTAGCAGAGGAGCTCAGCG-3′, which were specifically designed from the ICAM-1 promoter region (−346 to −24). PCR products were analyzed on ethidium bromide-stained agarose gels.
Neutrophil adhesion assay
Peripheral blood PMNs were isolated from whole venous blood by dextran sedimentation followed by density separation over Ficoll-Hypaque and hypotonic lysis. The PMNs were then re-suspended in Tyrode-HEPES buffer (128 mM NaCl, 2.7 mM KCl, 0.5 mM MgCl2, 0.36 mM NaH2PO4, 2 mM CaCl2, 12 mM NaHCO3, 10 mM HEPES, pH 7.4) and adjusted to 107 cells/ml. PMNs were used within 4 h after purification.
Adhesion of neutrophils to A549 cells was measured as described previously (Chen et al., 2001). Briefly, neutrophils were labeled with 2′,7′-bis (carboxyethyl)-5(6)-carboxyfluorescein (BCECF)/AM and added to the A549 monolayer. Non-adherent cells were removed by gentle washing with PBS, and the number of adherent cells was determined by measuring the fluorescence intensity using a Fusion (Packard Bioscience Co., Meriden, CT).
Statistical analysis
Data were analyzed with GraphPad Prism Program (GraphPad, San Diego, CA) and expressed as the mean ± SEM analyzed with a two-tailed Student's t-test at a P < 0.05 level of significance.
Results
Involvement of MAPKs and NF-κB pathways in IL-1β-induced ICAM-1 expression
In our previous study, IL-1β-induced ICAM-1 expression is mediated through MAPKs and NF-κB in A549 cells (Lin et al., 2005). In this study, as shown in Figure 1A, ICAM-1 expression was induced by IL-1β in a time-dependent manner. A maximal increase was achieved within 24 h. In addition, to compare the effect of IL-1 and IFN-γ for ICAM-1 expression, we have carefully performed ICAM-1 expression using concentrations of IFN-γ (0.01, 0.1, 1, and 10 ng/ml) and IL-1β (1.5, 3, 15, and 30 pg/ml) to treat A549 cells. At 10 ng/ml, IFN-γ can induce expression of ICAM-1 on A549 cells as well as IL-1β (3, 15, and 30 pg/ml) has evidently inductive effect, but at 1 ng/ml IFN-γ and 1.5 pg/ml IL-1β induced very little effect on ICAM-1 expression. These data suggested that the potency of IFN-γ-induced ICAM-1 expression is lower than that of IL-1β on A549 cells. The regulation of ICAM-1 expression occurred at transcription level through MAPKs and NF-κB pathways was further confirmed by gene luciferase activity assay. The promoter activity assays were performed using a human ICAM-1 promoter-luciferase construct, pIC339 (−339 to 0). Data in Figure 1B showed that IL-1β stimulated a maximal ICAM-1-luciferase activity within 5 h and declined to the basal level within 24 h. Moreover, IL-1β-stimulated ICAM-1 luciferase activity was significantly inhibited by pretreatment with U0126, SB202190, SP600125, and helenalin, respectively (Fig. 1C). The involvement of p42/p44 MAPK, p38, and JNK in IL-1β-stimulated ICAM-1 gene transcription was further confirmed by the results that co-transfection with dominant negative mutants of MEK1, ERK2, p38, and JNK, significantly attenuated ICAM-1 luciferase activity (Fig. 1D). These data confirmed that IL-1β-stimulated ICAM-1 luciferase gene activity occurred at a transcription level through activation of MAPKs and NF-κB pathways, consistent with our previous results revealed by Western blotting and RT-PCR analyses (Lin et al., 2005).
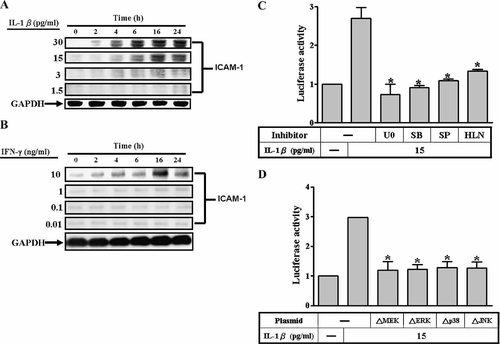
Involvement of MAPKs and NF-κB in IL-1β-induced ICAM-1 protein expression and promoter activity in A549 cells. A: Time dependence of IL-1β and IFN-γ-induced ICAM-1 expression, cells were cultured in serum-free medium at 37°C for 24 h and then treated with IL-1β and IFN-γ for various times. After incubation, Western blot analysis was performed, as described in Materials and Methods. Data are expressed as a representative of three independent experiments. For transcriptional regulation of ICAM-1 expression, A549 cells were transiently transfected with ICAM-1 reporter gene (pIC339-Luc) and then treated with IL-1β (15 pg/ml) for various times (B), and (C) pretreated with U0126 (10 µM), SB202190 (30 µM), SP600125 (10 µM), or helenalin (10 µM) for 1 h and then stimulated with IL-1β (15 pg/ml) for 5 h. D: Cells were cotransfected with plasmids encoding dominant negative mutants of MEK1, ERK2, p38, and JNK for 48 h, shifted to serum free DMEM/F-12 for 24 h, and then stimulated with IL-1β (15 pg/ml) for 5 h. Luciferase activity was determined in the cell lysates as described under Materials and Methods. Results are presented as means ± SEM from three separate experiments. *P < 0.05, as compared with the cells incubated with (B) the basal level or (C,D) cells exposed to IL-1β alone.
Involvement of Src/PDGFR/PI3K/Akt pathways in IL-1β-induced ICAM-1 expression
To further investigate the role of the Src/PDGFR/PI3K/Akt pathway in ICAM-1 gene transcription, A549 cells were transfected with ICAM-1 luciferase reporter gene (pIC-339), pretreated with inhibitors for 1 h, and then stimulated with IL-1β. As shown in Figure 2A, IL-1β-induced ICAM-1 promoter activation was inhibited by selective inhibitors of Src (PP1), PDGFR (AG1296), PI3-K (LY294002 and wortmannin), and Akt (SH-5), respectively. These results indicated the involvement of Src, PDGFR, and PI3K/Akt in IL-1β-induced ICAM-1 gene transcription.
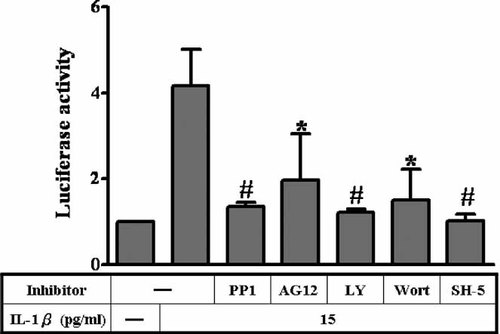
Inhibition of Src, PDGFR, PI3K, and Akt blocks IL-1β-induced ICAM-1 promoter activity. A549 cells transfected with ICAM-1 reporter gene (pIC339-Luc) were pretreated with PP1 (10 µM), AG1296 (10 µM), LY294002 (10 µM), wortmannin (10 µM), and SH-5 (10 µM) for 1 h before incubation with 15 pg/ml of IL-1β for 5 h. Luciferase activity was assayed as described in Materials and Methods. Data are presented as means ± SEM from three separate experiments. *P < 0.05; #P < 0.01, as compared with the cells exposed to IL-1β alone.
IL-1β-induced ICAM-1 expression mediates through Src phosphorylation
It has been demonstrated that TNF-α-induced ICAM-1 expression is mediated through the activation of Src-dependent pathways (Huang et al., 2003). To examine whether Src was also involved in IL-1β-induced ICAM-1 expression, pretreatment of A549 cells with a Src inhibitor (PP1) for 1 h prior to exposure to IL-1β for 5 h caused an attenuation of ICAM-1 expression in a concentration-dependent manner revealed by Western blotting analysis (Fig. 3A; P < 0.05, n = 3, as compared with the cells stimulated by IL-1β alone). To further ascertain that the activation of Src was required for IL-1β-induced ICAM-1 protein expression, A549 cells were transfected with a dominant negative mutant of Src, and then treated with IL-1β for 5 h. As shown in Figure 3B, transfection with dominant negative mutant of Src significantly attenuated ICAM-1 protein expression induced by IL-1β.
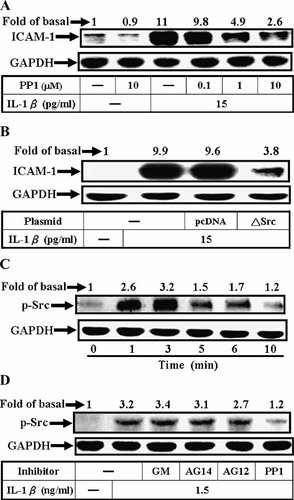
Effect of PP1 on IL-1β-induced ICAM-1 protein expression and Src phosphorylation in A549 cells. For ICAM-1 protein expression, the cells were grown to confluence on 12-well culture plates and shifted to serum free DMEM/F-12 for 24 h. The cells were pretreated with (A) various concentrations of PP1 for 1 h or (B) co-transfected with plasmids encoding dominant negative mutants of Src for 48 h, shifted to serum free DMEM/F-12 for 24 h, and then incubated with IL-1β (15 pg/ml) for 5 h. After incubation, Western blot analysis was performed, as described in Figure 1. Data are expressed as mean of three independent experiments. C: Time dependence of IL-1β-stimulated Src phosphorylation, cells were incubated with IL-1β (1.5 ng/ml) for various times. D: Inhibition of IL-1β-stimulated Src phosphorylation, A549 cells were pretreated with GM6001 (10 µM), AG1478 (10 µM), AG1296 (10 µM), or PP1 (10 µM) for 1 h and then stimulated with IL-1β (1.5 ng/ml) for 5 min. After incubation, Western blot analysis was performed, as described in Materials and Methods. Data are expressed as mean of three independent experiments.
Next, to determine whether Src phosphorylation was necessary for the IL-1β-induced ICAM-1 expression, activation of this kinase was assayed by blotting with an antibody specific for the phosphorylated, active form of Src. As shown in Figure 3C, IL-1β (1.5 ng/ml) stimulated a transient phosphorylation of Src in A549 cells. A maximal response was obtained within 3 min and declined to the basal level within 10 min. This IL-1β-stimulated Src phosphorylation was blocked by PP1, but not by AG1478, AG1296, or GM6001. These results indicated that IL-1β-stimulated Src phosphorylation was not mediated through EGFR, PDGFR, and MMPs or an up-stream component of growth factor receptors. These results suggested a link between transactivation of Src and induction of ICAM-1 expression by IL-1β in A549 cells.
IL-1β induces transactivation of PDGFR through Src phosphorylation and leads to ICAM-1 expression
Cell migration has been shown to be mediated through Src/PDGFR transactivation induced by extracellular stimuli (Qi et al., 1999). Hence, to further investigate whether PDGFR transactivation was involved in IL-1β-induced ICAM-1 expression, pretreatment of A549 cells with an inhibitor of PDGFR (AG1296) for 1 h prior to exposure to IL-1β for 5 h caused an attenuation of ICAM-1 expression in a concentration-dependent manner determined by Western blotting analysis (Fig. 4A; P < 0.05, n = 3, as compared with the cells stimulated by IL-1β alone). Next, we observed whether phosphorylation of PDGFR was required for IL-1β-induced responses, cells were treated with 1.5 ng/ml of IL-1β for the indicated times. Cell lysates were subjected to SDS–PAGE, transferred to membranes, and blotted with anti-phospho-PDGFR or GAPDH Ab. As shown in Figure 4B, IL-1β stimulated PDGFR phosphorylation in a time-dependent manner. A maximal response was obtained within 3–5 min and declined to the basal level within 10 min. This IL-1β-stimulated PDGFR phosphorylation was blocked by GM6001, PP1, and AG1296 (Fig. 4C). These results suggested that transactivation of PDGFR may be mediated through Src activation and involved in IL-1β-induced ICAM-1 protein expression in A549 cells.
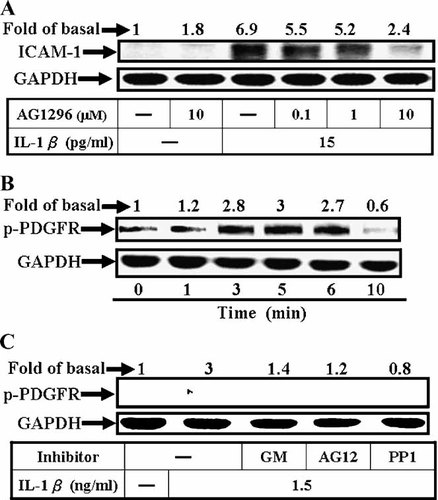
IL-1β-induced ICAM-1 is mediated through PDGFR phosphorylation in A549 cells. A: For ICAM-1 protein expression, cells were pretreated with various concentrations of AG1296 for 1 h and then stimulated with IL-1β (15 pg/ml) for 5 h. B: Time dependence of IL-1β-stimulated PDGFR phosphorylation, cells were incubated with IL-1β (1.5 ng/ml) for various times. C: PDGFR transactivation mediated through Src, cells were pretreated with GM6001 (10 µM), AG1296 (10 µM), and PP1 (10 µM) for 1 h and stimulated with IL-1β (1.5 ng/ml) for 5 min. After incubation, whole cell lysates were separated by a 10% SDS–PAGE, and immunoblotted with (A) anti-ICAM-1 or (B,C) antiphospho-PDGFR Ab. Data are expressed as mean of three independent experiments.
PI3K/Akt involves in IL-1β-induced ICAM-1 expression
Akt has been shown to be a downstream component of Src/RTK transactivation pathway (Buchanan et al., 2003). Transactivation of EGFR and PDGFR has been shown to regulate expression of various genes and proteins mediated via Src and Akt phosphorylation stimulated by IL-1β (Slomiany and Slomiany, 2004). Here, we presented evidence that Akt activation was involved in IL-1β-induced ICAM-1 protein expression. As shown in Figure 5A,B, pretreatment of A549 cells with PI3K inhibitors LY294002 and wortmannin for 1 h prior to exposure to IL-1β for 5 h caused an attenuation of ICAM-1 expression in a concentration-dependent manner by Western blotting analysis (P < 0.05, n = 3, as compared with the cells stimulated by IL-1β alone). To further ensure the involvement of PI3K and Akt in the expression of ICAM-1 induced by IL-1β, A549 cells were transfected with dominant negative mutants of p85 and Akt, and then treated with IL-1β for 5 h. As shown in Figure 5C, transfection with dominant negative mutants of p85 and Akt significantly decreased IL-1β-stimulated ICAM-1 expression. Taken together, these results suggested that PI3K/Akt was involved in IL-1β-induced ICAM-1 expression in A549 cells.
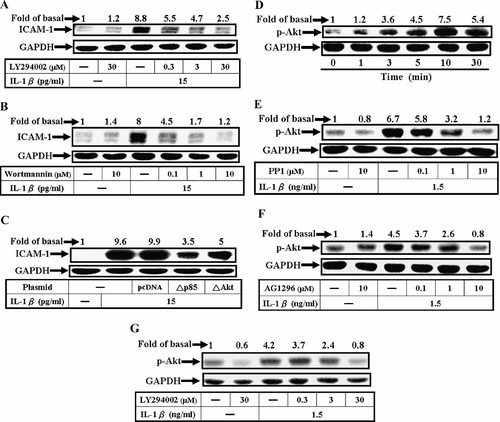
Effects of LY294002 and wortmannin on IL-1β-induced ICAM-1 protein expression and Akt phosphorylation in A549 cell. For ICAM-1 protein expression, the cells were pretreated with various concentrations of (A) LY294002 and (B) wortmannin for 1 h or (C) co-transfected with plasmids encoding dominant negative mutants of p85 and Akt for 48 h, shifted to serum free DMEM/F-12 for 24 h, and then incubated with IL-1β (15 pg/ml) for 5 h. After incubation, Western blot analysis was performed, as described in Figure 1. D: Time dependence of IL-1β-stimulated Akt phosphorylation, cells were incubated with IL-1β (1.5 ng/ml) for various times. E,F,G: Concentration-dependent inhibition of IL-1β-stimulated Akt phosphorylation, A549 cells were pretreated with various concentrations of PP1, AG1296, and LY294002 for 1 h and then stimulated with IL-1β (1.5 ng/ml) for 5 min. After incubation, Western blot analysis was performed, as described in Figure 1. Data are expressed as mean of three independent experiments.
Next, to corroborate the relationship between Akt phosphorylation and ICAM-1 expression, we determined Akt phosphorylation on residues Ser473 stimulated by IL-1β, which is a prerequisite for the catalytic activity of Akt (Downward, 1998). As shown in Figure 5D, IL-1β stimulated Akt phosphorylation in a time-dependent manner. A significant increase in Akt phosphorylation was observed within 3 min, peaked within 10 min, and sustained over 30 min. To confirm whether Akt phosphorylation was a downstream component of Src/PDGFR pathway, cells were pretreated with PP1, AG1296, or LY294002 for 1 h, and then treated with 1.5 ng/ml of IL-1β for 10 min. As shown in Figure 5E–G, Akt phosphorylation was inhibited by PP1, AG1296, or LY294002 in a concentration-dependent manner, indicating the requirement of Src and PDGFR transactivation for the Akt phosphorylation and ICAM-1 expression induced by IL-1β.
Src/PDGFR/PI3K/Akt is required for IL-1β-induced ICAM-1 mRNA expression
In the presence of IL-1β, analysis of RT-PCR showed that a maximal mRNA level of ICAM-1 was induced within 3–6 h (Fig. 6A). The possible regulation of ICAM-1 mRNA transcription by these kinases was also investigated using selective inhibitors. As shown in Figure 6B, pretreatment of A549 cells with inhibitors of c-Src (PP1), PDGFR (AG1296), and PI3K (LY294002) significantly attenuated IL-1β-induced ICAM-1 mRNA accumulation revealed by RT-PCR. These results further indicated that in A549 cells, regulation of ICAM-1 expression through activation of Src/PDGFR/PI3K/Akt pathway also occurred at the transcriptional level.
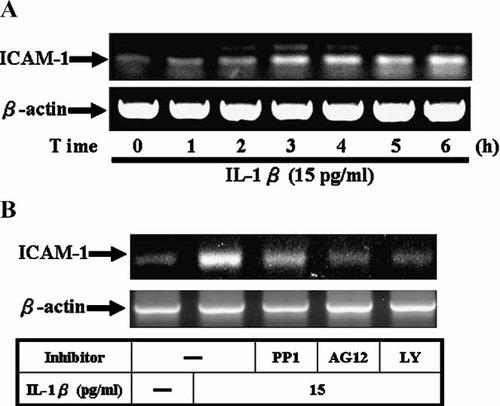
Involvement of Src, PDGFR, and PI3K/Akt in IL-1β-induced ICAM-1 mRNA expression in A549 cells. A: Time dependence of IL-1β-induced ICAM-1 mRNA expression, the cells were incubated with IL-1β (15 pg/ml) for the indicated times. B: Cells were pretreated with inhibitors of Src (PP1, 10 µM), PDGFR (AG1296, 10 µM), and PI3K (LY294002, 30 µM) for 1 h then the cells were incubated with IL-1β (15 pg/ml) for 5 h. The isolated RNA samples were analyzed by RT-PCR, using the primer specific for ICAM-1 and β-actin, as described in the text. Data are expressed of three independent experiments.
Inhibition of PI3K/Akt blocks IL-1β-induced recruitment of p300, acetylation of histone H3 on NF-κB binding site with ICAM-1 promoter
To investigate the role of the PI3K/Akt pathway in regulating the recruitment of p300 to the ICAM-1 promoter, a selective p300 inhibitor, curcumin (Balasubramanyam et al., 2004) was used. IL-1β-induced ICAM-1 expression was significantly inhibited by preincubation with curcumin (Fig. 7A), suggesting that p300 may involve in regulation of ICAM-1 gene expression. Furthermore, the in vivo association of p300 and acetyl-histione H3 to the ICAM-1 promoter induced by IL-1β was examined by ChIP. Chromatin was immunoprecipitated with anti-p300 and anti-H3 antibody, and the ICAM-1 promoter-enhancer region (−346 to −24) containing the NF-κB binding site for transcriptional activators was amplified by PCR. As shown in Figure 8B, in vivo binding of p300 and acetyl-histione H3 to the ICAM-1 promoter was increased after treatment with IL-1β in a time-dependent manner. Association of p300 and histone H3 with the ICAM-1 promoter was blocked by pretreatment with LY294002 (Fig. 7B). These data indicated that p300 and histone H3 were involved in IL-1β-induced ICAM-1 transcription and regulated by PI3K/Akt dependent pathway in A549 cells.
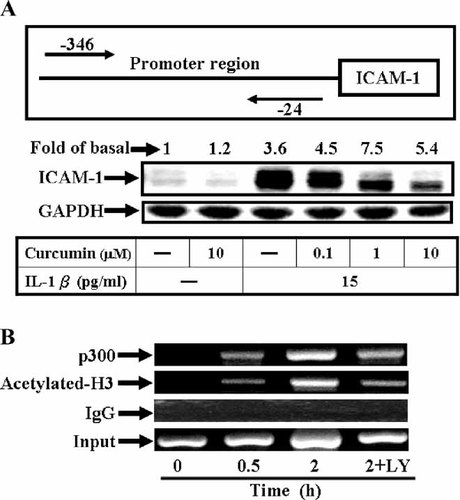
Effect of IL-1β on the in vivo DNA association of transcription factors at the ICAM-1 promoter in A549 cells. A: Cells were pretreated with curcumin for 1 h and then incubated with IL-1β for 5 h. After incubation, Western blot analysis was performed as described in Figure 1. Data are expressed as mean of three independent experiments. B: Confluent and serum-starved A549 cells in 100-mm dishes were incubated with IL-1β (1.5 ng/ml) for the indicated times. The in vivo protein-DNA complexes were cross-linked by formaldehyde treatment and chromatin pellets were extracted and sonicated. The associated ICAM-1 promoter DNA was amplified by PCR as described in Materials and Methods. The sequence of the human ICAM-1 promoter region (−346 to −24) amplified by PCR primer pairs was indicated by the arrows. An enrichment of the ICAM-1 promoter DNA is shown after PCR amplification of immunoprecipitates of p300- and histone H3-associated DNA from cells treated with IL-1β for the indicated times. The input represents PCR products from chromatin pellets prior to immunoprecipitation. The results are representatives of two independent experiments with similar results.
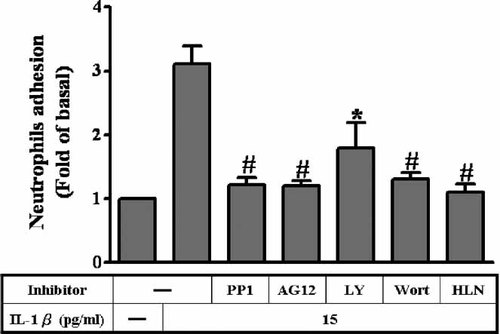
Inhibition of Src, PDGFR, and PI3K/Akt prevents IL-1β-induced PMN adhesion to A549 cells. Polymorphonuclear (PMN) cells, labeled with BCECF, were added to A549 cells pretreated with 10 µM PP1, 10 µM AG1296, 10 µM AG1478, 10 µM wortmannin, or 10 µM LY294002 for 1 h before incubation with IL-1 for 5 h, and culture was continued at 37°C for 1 h. Adhesion was measured as described in Materials and Methods. Data are expressed as mean ± SEM of three separate experiments. *P < 0.01; #P < 0.01, as compared with the cells incubated with IL-1β alone.
IL-1β-stimulated neutrophil adhesion is blocked by inhibitors of Src/PDGFR/PI3K/Akt on A549 Cells
Previous studies have demonstrated a potential role for IL-1β in increases adhesiveness between leukocytes and several cell types (Lin et al., 2005; Wang et al., 2005). In the present study, to test the adhesive activity between neutrophils and A549 cells, we isolated neutrophils from human blood and labeled with 2′,7′-bis (carboxyethyl)-5(6)-carboxyfluorescein (BCECF) and added to the A549 monolayer prior exposure to IL-1β. In parallel with the inhibition of ICAM-1 expression, the A549 cells were pretreated with 10 µM PP1, 10 µM AG1296, 10 µM wortmannin, 10 µM LY294002, and 10 µM helenalin and then incubation with 15 pg/ml IL-1β. As shown in Figure 8, the amount of neutrophils adhesion to A549 cell monolayer was significantly increased (approximate three folds) by incubation with 15 pg/ml IL-1β for 5 h (P < 0.05, n = 3, as compared with the basal level). The specific inhibitors for Src, PGGFR, PI3K, and Akt can block IL-1β-stimulated neutrophil adhesion. In addition, neutrophils adhesion to IL-1β-stimulated A549 cell monolayers was also significantly inhibited by preincubation with antibody to ICAM-1 (data not shown). These results further supported that in A549 cells, up-regulation of ICAM-1 expression enhanced adhesion between A549 cells and PMNs through Src/PDGFR/PI3K/Akt pathway.
Discussion
These results demonstrate that Akt plays an important role in mediating the IL-1β-induced ICAM-1 expression in A549 through the downstream activation of p300 and histone H3 acetylation. In our previous study, expression of ICAM-1 at the protein and mRNA levels was also increased in a time-dependent manner, which was blocked by pretreatment with selective inhibitors for MAPKs and NF-κB (Lin et al., 2005). This mechanism was supported by transfection of A549 cells with an ICAM-1 luciferase reporter gene. We showed the first, IL-1β stimulated ICAM-1-luciferase activity within 3 h and sustained for over 5 h (Fig. 1A). Moreover, activation of ICAM-1 luciferase activity by IL-1β was significantly inhibited by pretreatment with helenalin, U0126, SB202190, SP600125, PP1, AG1296, LY294002, and wortmannin (Figs. 1B and 2A). Co-transfection with dominant negative plasmids of ERK, p38, JNK, and MEK also decreased ICAM-1-luciferase activity in response to IL-1β. In the functional assay, adhesion of neutrophils to A549 cells was blocked in parallel by the inhibition of the phosphorylation of Src, PDGFR, Akt, and NF-κB (Fig. 8). These results support the notion that in A549 cells, IL-1β-induced ICAM-1 expression is mediated not only by MAPKs pathway, but also by Src/PDGFR/PI3K/Akt and NF-κB signaling pathways.
It has been demonstrated that Src family kinase plays a key role in the transduction of signals by GPCRs and growth factor receptors, which are involved in cell migration and proliferation (Krymskaya et al., 2004). In addition, Src also plays a major role in regulation of adhesion molecule expression in human airway epithelial cells treated with cytokines (Chang et al., 2004). In A549 cells, IL-1β-induced ICAM-1 expression was significantly blocked by a Src inhibitor PP1 and transfection with dominant negative plasmid, Src (K295M). Moreover, IL-1β had been shown to induce phosphorylation of Src which was also inhibited by PP1, not by inhibitors of MMP (GM6001), EGFR (AG1478), and PDGFR (AG1296). These results were consistent with our previous study that in rat vascular smooth muscle cells, Src kinases phosphorylation is required for receptor tyrosine kinase, Akt, and p42/p44 MAPK activation in response to BK, since treatment with a Src kinase inhibitor PP1 attenuated the phosphorylation of these signaling proteins (Yang et al., 2005).
Here, we have also observed that IL-1β-dependent activation of PDGFR and Akt was mediated through Src. A Src inhibitor PP1 and PDGFR kinase inhibitor AG1296 abrogated IL-1β-induced ICAM-1 expression in A549 cells. Activation of PI3K appears to occur via phosphorylation of tyrosine residues in the Src homology 2 domain of p85. In this study, IL-1β-stimulated phosphorylation of Akt was blocked by PP1 and AG1296. Furthermore, inhibition of PI3-K by LY294002 also attenuated IL-1β-induced Akt phosphorylation, but had no effect on Src and PDGFR phosphorylation (data not shown), suggesting that Src and PDGFR may be upstream components of PI3-K/Akt in regulation of ICAM-1 expression. The involvement of PI3K/Akt in ICAM-1 expression was supported by that transfection of A549 cells with dominant negative plasmids of p85 and Akt significantly attenuated IL-1β-induced ICAM-1 expression.
Indeed, the evidence has demonstrated that Akt is activated and translocated into the nucleus in response to insulin growth factor and cytokines, such as TNF-α (Andjelkovic et al., 1997; Huang and Chen, 2005). Nuclear Akt associates with p300 may lead to enhance HAT activity. The HAT such as p300 and CBP are phosphoproteins which have been recognized as key molecules involved in the communication between the transcription factors and the transcriptional machinery, thus appearing to be important in the gene regulation network (Chan and La Thangue, 2001). Histone acetylation is attributed to the Akt-enhanced intrinsic HAT activity of p300 and its association with another HAT, p/CAF (Goodman and Smolik, 2000). Acetylation of histone results in charge neutralization of conserved, often invariant, lysine residues located in amino-terminal domains of the histones. These domains are often referred to as tails because they protrude out from the nucleosome core. Changes in the charge of the histone tails are hypothesized to weaken histone-DNA or histone–histone contacts. Any or all of these changes can affect the structure of individual nucleosomes as well as higher-order folding, leading to a more open and permissive chromatin environment for binding of transcriptional factor, such as NF-κB (Roth et al., 2001). The C-terminal part of p300 (1,829–1,834) contains a consensus motif RXRXXpS/T, where X is any amino acid, R is arginine, and pS/T is phosphorylated serine or threonine, preferred by Akt has been characterized (Alessi et al., 1996). Akt has also been demonstrated to phosphorylate p300/CBP within their C-terminal parts and to increase their HAT and/or transcriptional activities. Using ChIP assays, the data showed that stimulation of IL-1β for 2 h increased the recruitment of p300 to the ICAM-1 promoter and the increase in HAT activity, which results in the acetylation of histone H3 on the ICAM-1 promoter. These data provide evidence that the synergic effect of HAT activity induced by p300 is Akt-dependent and is essential for ICAM-1 gene transcription. These effects were attenuated by pretreatment with LY294002. Thus, activation of Akt by Src/PDGFR signaling may eventually lead to enhance HAT activity induced by p300 and ICAM-1 gene expression.
In addition, our previous study has demonstrated that MAPKs and NF-κB play an important role in expression of ICAM-1 gene. In the present study, L-1β-induced ICAM-1 expression was further confirmed by transfection with ICAM-1 luciferase reporter gene. These data showed that IL-1β-induced ICAM-1 luciferase activity was inhibited by co-transfecting dominant negative plasmids including MEK, ERK, p38, and JNK, as well as their selective inhibitors of MEK1/2 (UO126), p38 (SB202190), JNK (SP600125), and NF-κB (helenalin), respectively. These data suggested that activation of MAPKs and NF-κB pathways regulated ICAM-1 expression at a transcription level. However, whether the activation of these three MAPKs led to NF-κB translocation and finally resulting in ICAM-1 expression through promotion of p300/CBP activating remains unknown in A549 cells. Previous studies have shown that p300/CBP can be phosphorylated by three of MAPKs and regulates gene expression (Ait-Si-Ali et al., 1999; Darieva et al., 2004; Poizat et al., 2005). Furthermore, p300/CBP has been reported to increase the transcriptional activity of NF-κB through acetylation (Deng et al., 2003; Chen et al., 2005) resulting in the respective prosurvival and proapoptotic effects. The phosphorylation and regulation of p300 by these kinases probably provide a novel mechanism determining cell survival and proliferation.
In summary, this study demonstrates that in A549, the mechanisms underlying IL-1β receptor-mediated through transactivation of Src/PDGFR/PI3K and Akt as well as p300 were required for expression of ICAM-1 (Scheme 1). This regulatory mechanism may have important implications in the gene transcription related to the pathogenesis of airway inflammatory diseases.
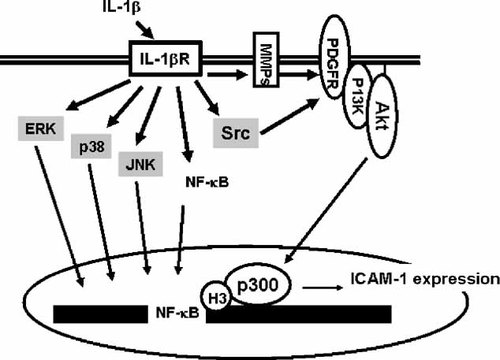
Schematic representation of the signaling pathways involved in IL-1β-induced ICAM-1 expression in human pulmonary epithelial cells. IL-1β receptor and activates at least three independent pathways involved in Src/EGFR/PDGFR/PI3K/Akt, MAPKs and NF-κB activation. Phosphorylation of Akt was translocated into nucleus and activated p300. Finally trigger chromatin remodeling and promote NF-κB binding to ICAM-1 promoter binding site.
Acknowledgements
The authors appreciate Dr. K. L. Guan (Department of Biological Chemistry, University of Michigan, MI, USA), Dr. M. H. Cobb (Department of Pharmacology, University of Texas Southwestern Medical Center), Dr. R. D. Yeh (Department of Pharmacology, University of Illinois at Chicago), Dr. C. C. Chen (Department of Pharmacology, National Taiwan University), Dr. J. Han (The Scripps Research Institute, La Jolla, CA, USA), and Dr. P. T. van der Saag (Hubrecht Laboratory, Utrecht, the Netherlands) for providing dominant negative mutants of MEK, ERK2 (ERK2 K52R), p85, Akt, Src, JNK, p38, and ICAM-1(pIC-339/0)/firefly luciferase (Luc) construct, respectively.




