A novel de novo dominant mutation of NOTCH1 gene in an Iranian family with non-syndromic congenital heart disease
Funding information
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abstract
Background
Congenital heart disease (CHD) is the most common birth defect which can arises from different genetic defects. The genetic heterogeneity of this disease leads to restricted success in candidate genes screening method. Emerging approaches such as next-generation sequencing (NGS)-based genetic analysis might provide a better understating of CHD etiology in the patients who are left undiagnosed. To this aim, in this study, we survived the causes of CHD in an Iranian family who was consanguineous and had two affected children.
Methods
Affected individuals of this family were checked previously by PCR-direct sequencing for six candidate genes (NKX2-5, ZIC3, NODAL, FOXH1, GJA1, GATA4) and had not revealed any reported CHD causative mutations. Whole-exome sequencing (WES) was performed on this family probond to determine the underlying cause of CHD, and the identified variants were confirmed and segregated by Sanger sequencing.
Results
We identified one heterozygous missense mutation, c.T6797C (p.Phe2266Ser), in the NOTCH1 gene, which seems to be the most probably disease causing of this family patients. This mutation was found to be novel and not reported on 1000 Genomes Project, dbSNP, and ExAC.
Conclusion
Worldwide, mutations in NOTCH1 gene are considered as one of the most known causes of CHD. The found NOTCH1 variant in this family affected individuals was the first report from Iran. Yet again, this result indicates the importance of NOTCH1 screening in CHD patients.
1 INTRODUCTION
Congenital heart disease (CHD) as a heart structural abnormality affects 8/1000 live births in worldwide.1 CHD indicates a variety of heart defects including outflow tract defects, valve defects, and septal defects 2 such as ventricular septal defect (VSD) which is the most common CHD and observed in girls and boys with similar frequency, patent ductus arteriosus (PDA) considers for 10% of CHDs and often has unknown reason 3 and atrioventricular septal defect (AVSD) constitutes more than 7.4% of CHDs, it is result from endocardial cushion defects.4
Causes of CHD are classified into genetic, epigenetic, and environmental categories and in about 90% of cases, no determined cause can be described.5 The majority of CHDs occur just as a heart defect without other organs anomaly, however, may some syndrome co-occurrence with CHD. Therefore, CHDs can be considered syndromic or non-syndromic. The CHD risk for a kindred offspring mostly depends on the CHD etiology in the parent. Mutations in numerous genes which are involved in heart development, that is, signal transduction, transcriptional regulation, and encoding cardiac proteins, lead to non-syndromic CHDs. The NOTCH conduction pathway is a critical process which plays significant role in left-right axis subdividing.5
NOTCH1 gene is located on chromosome 9q34.3 and consist 34 exons. It encodes 300kDa protein, which functions as a transmembrane receptor.6 NOTCH1 protein regulates several critical process, such as vasculogenesis and cardiac development.7 Mutations in NOTCH1 gene are reported in a range of CHD, comprising bicuspid aortic valve (BAV),8 aortic stenosis (AS),9 hypoplastic left heart syndrome (HLHS),10 coarctation of the aorta (COA),5 and tetralogy of fallot (TOF).11
At present, based on Human Gene Mutation Database (HGMD) (www.hgmd.cf.ac.uk), 30 mutations causing CHD have been introduced in the NOTCH1 gene including 21 missenses/nonsenses, three splicings, two small deletions, and 4 gross deletions. To our knowledge, no NOTCH1 mutations were observed in Iranian CHD patients. Furthermore, there are no VSD, PDA, and AVSD clinical reports of NOTCH1 gene mutations in worldwide. In this study, we report a novel de novo germline mutation in the NOTCH1 gene that was detected by whole-exome sequencing (WES), associated with VSD, PDA, and AVSD in an Iranian family.
2 MATERIAL AND METHODS
2.1 Patient
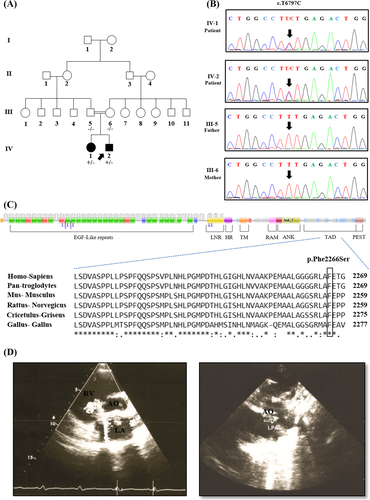
An Iranian healthy consanguineous couple, 30-year-old mother and a 33-year-old father, who had two affected children was referred to the Rajaie Cardiovascular, Medical and Research Center, Tehran, Iran for genetic counseling (Figure 1A). No syndromic characterizes were identified in their medical history as well as in clinical investigations. The diagnosis of CHDs, 8-year-old girl with VSD and 6-month-old boy with PDA and AVSD (Figure 1D), was confirmed by echocardiogram. With informed consent, blood samples were collected from individuals IV-1 and IV-2 and their biological parents (III-5 and III-6). A diagnostic algorithm starting with Karyotyping followed by a PCR-direct Sequencing for six candidate genes (NKX2-5, ZIC3, NODAL, FOXH1, GJA1, GATA4), MLPA (multiplex ligation-dependent probe amplification) and Array-CGH (array comparative genomic hybridization) previously was performed for this family by our team had not revealed any reported CHD causative mutations. This study was performed due to the Declaration of Helsinki Principles and was reviewed by the ethics committee of Rajaie Cardiovascular, Medical and Research Center.
2.2 DNA extraction and Whole-exome sequencing
DNA was extracted from blood samples using standard salting-out method.12 DNA samples were assessed by a NanoDrop (Thermo Fisher Scientific) and agarose gel electrophoresis to confirm the quality and quantity of the DNAs.
Exome sequencing was performed only on the proband, patient IV-2 (Figure 1A) at Macrogen. Briefly, 10 ng of genomic DNA was used for WES (library construction, exome capture and reads sequencing) by SureSelect XT Library Prep Kit on an Illumina HiSeq 4000 according to the manufacturer's protocol (Illumina). The generated reads were aligned to the human reference genome (NCBI GRCh37/hg 19 version) with Burrows-Wheeler Aligner (BWA) alignment software (http://bio-bwa.sourceforge.net/).13 Genome Analysis Toolkit (GATK) (https://www.broadinstitute.org/gatk/) 14 was used to call sequence variants. Genetic variants annotation were carried out with the ANNOVAR (http://annovar.openbioinformatics.org/) 15 by consideration of splice site, intronic, exonic, 5´ UTR, 3´ UTR, intergenic, upstream, or downstream locations and then frame shift, synonymous, non-synonymous, insertion/deletion or stop exonic functions. To make possible detection of de novo variants, we considered that disease-causing variants would be absent in the public available populations. Therefore, the variants were filtered against 1000 Genomes Project, the Single Nucleotide Polymorphism Database (dbSNP) and Exome Aggregation Consortium (ExAC) with respect to minor allele frequency (MAF) < %1. To analyses of non-synonymous variants, we used the result of ANNOVAR based on prediction tools such as mutation taster,16 sorting intolerant from tolerant (SIFT),17 polymorphism phenotyping v2 (PolyPhen-2) 18 and combined annotation dependent depletion (CADD).19 The variants were selected if they were recognized to be damaging by two of the prediction tools. Nucleotide numbering was according NCBI Gene Bank cDNA Accession Number NM_017617. ACMG Standards were used for the sequence variants interpretation.20
2.3 Segregation analyses
Candidate variant was validated and segregated in family members by standard Sanger sequencing. One PCR primer pair were designed using Primer3 v. 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/).21 To amplify of the variant, polymerase chain reaction (PCR) was performed on a SimpliAmp™ Thermal Cycler (Thermo Fisher Scientific) considering 100 ng DNA, 10 PM primers (Forward primer, AAGGCACGGAGGAAGAAGTC and Reverse primer: AGGGTTGTATTGGTTCGGC) 1.5 mmol/L MgCl2, 200 mmol/L dNTP, and 1 U of Taq DNA polymerase (Amplicon). PCR was ordered as follows: incubation at 95°C for 5 minutes and then 35 amplification cycles (40 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C). The confirmed PCR products by gel electrophoresis were directly Sanger sequenced on an ABI Sequencer 3500XL PE (Applied Bio Systems) and were analyzed using Codon Code Aligner v. 7.1.2 (https://www.codoncode.com/aligner/). When the variant was confirmed, all family members (healthy/patient) were surveyed to define variant segregation.
2.4 Protein analyses
The NOTCH1 protein domains have been obtained by Pfam v. 32.0 (https://pfam.xfam.org/)22 with Uniprot code P46531 (Figure 1C/Up). We also used CLUSTALW (https://www.genome.jp/tools-bin/clustalw)23 for multiple protein sequence alignment in the human as compared with other species (Figure 1C/Down).
3 RESULTS
The CHD of the family children, 8-year-old boy with VSD and 6-month-old girl with PDA and AVSD (Figure 1D) were diagnosed according to clinical examinations, that is, angiography, echocardiography, and MRI, by expert cardiologist. WES was performed on patient IV-2 DNA sample. After WES results filtering, nine candidate variants including seven dominants and two recessives were determined in the probond. Among these, only a missense variant, c.T6797C (p.Phe2266Ser), which identified in NOTCH1 gene, was found as the most probably disease reason in this family affected individuals. This heterozygous variant was not present in 1000 Genomes Project, dbSNP and ExAC.
Sanger sequencing of the patient and her family members DNA samples validated this mutation. Her affected brother (IV-1) carried this substitution; however, both their parents (III-5 and III-6) had normal sequence (Figure 1B). This suggests that a de novo mutation would be occurred in one of the parent's germline which has not detected by somatic-line Sanger sequencing but inherited to the children.
The novel de novo mutation c.T6797C was located within the exon 34 of the NOTCH1 gene. In wild type NOTCH1, the TTT nucleotides at c.6796-6798 position encode phenylalanine at p.2266. The c.6797T > C substitution resulting in Phe2266 changes to serine. The pathogenicity effect of this mutation was confirmed by ANNOVAR prediction tools (Mutation taster, SIFT, PolyPhen-2, and CADD). Furthermore, alignment of the targeted region has indicated this position is highly conserve among various species (Figure 1C/Down).
4 DISCUSSION
CHD is a complex disease which often has poorly understood reason. Approximately 8% of CHDs are associated with single genes mutations, 2% with environmental agents, and 90% have multifactorial etiology.5 In the last decades, several single gene variants have been determined in patients with non-syndromic CHDs, including NKX2-5, GATA4, GATA6, MYH6, TBX5, and TBX20.24 Although probable gene sequencing has detected multiple disease-causing mutations in many disorders, it has had restricted achievement in CHD, as described about our cases which were screened for six candidate genes (NKX2-5, ZIC3, NODAL, FOXH1, GJA1, GATA4) in previous study of our team and had not revealed any CHD causative mutations, likely because of genetic heterogeneity that numerous genes are involving in heart development. New approaches such as NGS can provided CHD genetic etiology.25
Our study indicates the first novel de novo germline mutation identified by WES in an Iranian family with different types of CHD. Although we surveyed nine likely pathogenic variants in the family members but none of them were completely segregated except one, c.T6797C in the NOTCH1 gene, and this illustrates the complexity of the CHD causing identification. According to the previous studies 11, 26-28 and our finding, we think the NOTCH1 c.T6797C (p.Phe2266Ser) variant have a significant role in the CHD of this family. The relation of NOTCH1 with CHD is consistent with NOTCH1 roles during cardiac development. NOTCH1 is in the endocardium and NOTCH1 knockout mice harbors abnormality in the ventricle and cardiomyocyte.29 NOTCH signaling leads to epithelial-to-mesenchymal transduction which is critical for valvulogenesis. The mutations of this pathway proteins, NOTCH1 and NOTCH2, decrease ligand-induced signaling and cause aortic valve malformations.9 In a study by Krebs et al,30 on NOTCH mutant mouse was demonstrated that NOTCH pathway has a major role in the LR asymmetry establishment by expression regulation of the NODAL gene.
NOTCH1 protein consists eight structural domains, including epidermal growth factor (EGF), Lin/Notch repeat (LNR), heterodimerization domain (HD), transmembrane domain (TD), RBPJ-associated molecule domain (RAM), ankyrin repeat (ANK), transactivation domain (TAD), and PEST domain (PEST) (Figure 1C/Up). Phenylalanine 2266 is lined in a TAD domain of NOTCH1 which is from amino acids 2193-2396 and induces autonomous transcription. The phenylalanine residue at p.2266 and around amino acids are highly conserved among different species (Figure 1C/Down). In our study, the identified mutation was located at the TAD domain of NOTCH1, thus impairs the function of this domain in target gene transcription, such as the Hey1 and Hey2 genes which are required for the canal myocardial boundary.31 Gerhardt et al generated knockout mice lacking NOTCH1 TAD to investigate the role of this domain, their functional assays displayed the importance of the TAD in mammalian cardiac development.32
NOTCH1 variants have been observed in several types of CHD including BAV,33 AS, HLHS, COA, TOF,34 left ventricular outflow tract obstructive (LVOTO),35 and pulmonary stenosis (PS).36 In our pedigree, we found a patient with VSD (IV-1) and other with PDA and AVSD (IV-2). Autosomal dominant (AD) inheritance and variable expressivity which were observed in this pedigree could lead to different phenotypes of the offsprings. As seen in different types of CHD, the phenotype for a specific mutation may be not the same.24
De novo variants are an important reason of early-onset genetic disease such as CHD, hearing loss.37 The identified NOTCH1 variant was found to be novel de novo which not reported on 1000 Genomes Project, dbSNP, and ExAC. Given that some of NOTCH1 mutations were also identified in healthy parents and considers as rare variants with reduced penetrance, presenting of the novel pathogenic variant in both affected individuals of a kindred and absent in healthy parents, highlights the hypothesis that dominant NOTCH1 c.T6797C variant has occurred in one of the parent's germ line and transmitted to children, this happening is probably more from father side according to the study of Kong et al which identified a de novo mutation in both sibling of a family but not the parents, they had believed de novo mutations in men different sperms are not completely independents.38 Mutational hotspots as a possible source of de novo mutations and founder effect which may lead to population-specific variants are important to consider for primary care plans.39, 40
It should be noted as a limitation of the present study, WES will miss regulatory/intronic variants which may have role in the phenotype modifying, as it was observed in the ERBB4 41 and ISL142 genes which these loci non-coding regions were associated with CHD susceptibility.
In conclusion, we report a family with CHD with a novel de novo germline mutation. Regarding the CHD causing mechanisms are largely obscure and the variant spectrum of non-syndromic CHD, we suggest that WES approaches could identify novel sequence changes to improve our understanding about CHD etiology. However, the importance of bioinformatics challenges and family analyses information should be considered in this subject.
ACKNOWLEDGMENTS
Special acknowledgments to the family that let us to document their story to improve our realization of the condition. This research provided by Rajaie Cardiovascular, Medical and Research Center, Tehran, Iran approved by RHC Ethics Committee (RHC.AC.IR.REC.1395.46; 24 December 2016) and Zanjan University of Medical Science, Zanjan, Iran approved by ZUMS Ethics Committee (ZUMS.REC.1396.145; 21 June 2017).
ETHICAL CONSIDERATIONS
Informed consent has been obtained by the authors.




