Differential Histone Modification Status of Spermatozoa in Relation to Fertility of Buffalo Bulls
ABSTRACT
In this study genome-wide di-methylated H3K4 (H3K4me2) and tri-methylated H3K27 (H3K27me3) modification profiles were analyzed in spermatozoa of buffalo bulls having wide fertility differences. The custom designed 4 × 180 K buffalo (Bubalus bubalis) ChIP-on-chip array was fabricated by employing array-based sequential hybridization using bovine and buffalo genomic DNA for comparative hybridization. The buffalo specific array developed had 177,440 features assembled from Coding sequences, Promoter and CpG regions comprising 2967 unique genes. A total of 84 genes for H3K4me2 and 80 genes for H3K27me3 were found differentially enriched in mature sperm of high and sub-fertile buffalo bulls. Gene Ontology analysis of these genes revealed their association with different cellular functions and biological processes. Genes identified as differentially enriched between high and sub-fertile bulls were found to be involved in the processes of germ cell development, spermatogenesis and embryonic development. This study presents the first genome-wide H3K4me2 and H3K27me3 profiling of buffalo bull sperm. Results provide a list of specific genes which could be made responsible for differential bull fertility. J. Cell. Biochem. 116: 743–753, 2015. © 2014 Wiley Periodicals, Inc.
A sperm carries haploid paternal genome to the oocyte on fertilization where the quality of DNA it carries play determining role for successful fertilization, orderly embryonic cleavage and successful implantation of embryos [Spano et al., 2000; Miller et al., 2005; Peddinti et al., 2008; Feugang et al., 2009, 2010]. Male factor infertility is considered as one of the primary concerns for implementing any planned animal improvement strategy [Mahmoud and Nawito, 2012]. Several genetic and epigenetic factors like aneuploidy, Y chromosome microdeletion, altered DNA methylation, chromatin modifications and others [Miller and Ostermeier, 2006; Ferlin et al., 2007; Krausz and Giachini, 2007; Miller et al., 2010] have been described as responsible elements in causing male infertility, however, the molecular mechanisms involved remains largely obscure. In the course of development, most of the epigenetic factors are modulated during the process of gamete formation [Reik et al., 2001; Carrell and Hammoud, 2010]. During spermatogenesis, the genome undergoes major changes including replacement of nucleosomal histones with arginine rich small basic proteins, protamine, and global remodeling of its epigenetic markings affecting elongation and compaction of sperm nucleus. The histone to protamine replacement causes greater than tenfold compaction of sperm chromatin which is required for effective delivery of the paternal genome to the oocytes [Balhorn et al., 2000; Miller et al., 2010]. Replacement of histone by protamine is however, not a complete process and it has been observed that 1% of the total DNA in mouse and about 15% in human still remains nucleosome bound [Tanphaichitr et al., 1978; Gatewood et al., 1990; Bench et al., 1996]. The rarely retained nucleosomes in sperm consist of either canonical or histone variant protein like H3K4me2, H3K4me3, H3K27me3 etc., including a testes-specific histone H2B (TH2B) [Kimmins and Sassone-Corsi, 2005] which are subjected to various modifications, including methylation, acetylation, phosphorylation, ubiquitination, and ribosylation to facilitate chromatin access in a time and space specific manner [Bannister and Kouzarides, 2011; Petty and Pillus, 2013]. Functionally, these modifications facilitate the protein-DNA and protein–protein interactions and regulate the development process through modifying the interaction of transcriptional regulators with chromatin [Strahl and Allis, 2000; Lee et al., 2010]. Further, it has been evidenced that the retained histones and its modified forms in sperm genome are significantly enriched at loci of developmental importance, including imprinted gene clusters, microRNA clusters and the promoters of developmentally important genes [Hammoud et al., 2009]. Also, these retained histones in sperm genome have been often found bivalently modified with both activating and silencing marks similar to stem cells [Bernstein et al., 2006; Brykczynska et al., 2010] which imparts special capability to sperm genome to play regulatory roles in the developing embryos [Hammoud et al., 2009; Brykczynska et al., 2010].
In the present study, we hypothesized that a genome wide profile of H3K4me2 and H3K27me3 histone modification patterns in buffalo sperm will be helpful to understand the reason behind observed variations in fertility of breeding bulls and eventual discovery of molecular biomarkers for male fertility.
MATERIALS AND METHODS
All chemicals were purchased from Sigma–Aldrich (St. Louis, MO), unless otherwise stated.
CLASSIFICATION OF BULLS IN HF AND SF GROUPS BASED ON THEIR CR RECORD AND IVF DATA
Ten bulls, five each in high-fertile (HF) and sub-fertile (SF) groups out of thirty eight bulls considered in this study were selected on the basis of conception rate (CR) and in vitro fertilization (IVF) data. These 38 bulls were used under multi herd progeny testing program in India. All bulls were reared under uniform feeding and management schedule and were inducted into progeny testing programme after evaluation of their breeding soundness, semen quality and freezability parameters. Frozen semen samples (0.25 ml straws) from these bulls were assigned randomly for breeding buffalo cows in the buffalo herds maintained with uniform feeding and management schedule. All these 38 bulls were having more than 50 insemination records over a period of 12 years (2000–2011). The SF bulls had a CR in the range of 26–36% whereas the CR of HF bulls were in the range of 54–58%. The cryopreserved semen samples of the identified bulls spread over random ejaculates were obtained from respective frozen semen stations. IVF was done using optimized in vitro oocytes maturation (IVM) and in vitro fertilization (IVF) procedure [Verma et al., 2012] with the difference that in vitro culture (IVC) of embryos was carried out in modified Charles Rosenkrans 2 amino acid (mCR2aa) based in vitro culture medium supplemented with 1% (v/v) minimal essential media (MEM) non-essential amino acids, 2% (v/v) MEM essential amino acids, 0.14 mg/ml L-glutamine, 1.5 mM glucose, 0.36 mg/ml sodium lactate, 0.036 mg/ml sodium pyruvate, 0.8% (w/v) BSA and 50 μg/ml gentamicin. IVC medium was replaced after 72 h of culture with replacement medium (IVC medium with 10% [v/v] FBS). A second media change of replacement medium was done on the 5th day of culture. Embryos were cultured up to 7 days, and different cleavage stages (2, 4, 8, 16, morula and blastocyst) were recorded at 36, 48, 60, 84, 120, and 168 h post-insemination, respectively. All experiments were repeated four times.
DESIGNING OF BUFFALO SPECIFIC ChIP-on-chip ARRAY
Since the fully annotated genome sequence is not available for buffalo, we employed the microarray based comparative genome hybridization (aCGH) principle to develop a buffalo specific ChIP-on-Chip array. The array was designed in two stages. In the first stage, a 1 M bovine array was designed using bovine genome (Build bosTau4) sequence data retrieved from UCSC Genome Browser (http://genome.ucsc.edu). This 1 M bovine array comprised of 60-mer oligonucleotide probes designed from the coding sequences (CDS), promoters, CpG rich regions and other genomic regions of bovine genome. The criteria for designing the probes were: GC% (35–55), melting temperature (65–90°C), devoid of palindromes, repeat regions and mono nucleotide repeats using Genotypic_Probe_Parser.pl (Genotypic Technology, Bangalore, India). Thus, the designed bovine 1 M array (AMADID—034986) consisted of 971,042 probes and 2974 Agilent control spots totaling to 974,016 probes (Table I). Bovine and Bubaline genomic DNA samples labeled respectively with Cy3 and Cy5 were hybridized to this array using aCGH protocol recommended by Agilent technologies. The probes which hybridized equally well with bovine and buffalo genomic DNA samples and exhibited significantly higher signal intensity (P < 0.05), at least twofold higher as compared to background were selected for further buffalo specific ChIP-on-Chip array design. Based on this criteria, a total of 177,440 probes from CDS, promoter and CpG rich regions representing 2967 unique genes were selected from 1 M bovine array to design the buffalo specific ChIP-on-Chip array in 4 × 180–K format (AMADID: 040096).
| Target Region | Avg tiling (bp) | Total no. of probes designed (1M array) | No. of probes on 180 K array | No. of genes | No. of unique genes |
|---|---|---|---|---|---|
| CDS | 20 | 262077 | 3310 | 1568 | 2967 |
| CPG | 4 | 242761 | 160708 | 0 | |
| Promoter | 45 | 242760 | 13422 | 1754 | |
| Other regions | 150 | 223444 | 0 | 0 |
CHROMATIN IMMUNOPRECIPITATION ASSAY
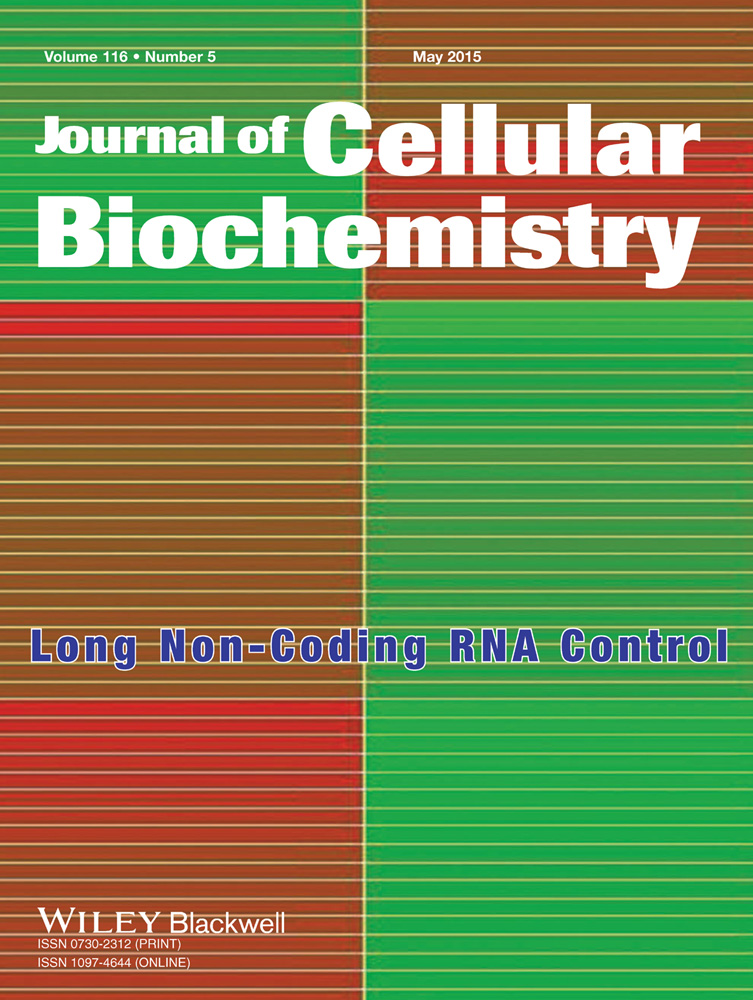
Before going for chromatin immunoprecipitation the semen samples (6–7 × 107 sperm) were treated with a hypotonic buffer; somatic cell lysis buffer containing 0.5% (v/v) Triton-X and 0.1% (w/v) SDS [Ostermeier et al., 2002; Goodrich et al., 2007] for 10 min at 4°C. Absence of contaminating somatic cells was confirmed by visual inspection of each sample under ×400 magnification using an Olympus BX51 microscope (Olympus America, Center Valley, PA) (Fig. 1a) and amplifying CD4 and CD45 genes from the somatic cell lysed sperm RNA preparation. The negative amplification of these genes confirmed somatic cell contamination free RNA preparation while positive amplification of PRM1 confirmed the presence of sperm RNA (Fig. 1b). Primer sequences for amplification of PRM1, CD4, and CD45 are given in Table II. Subsequently, the somatic cell lysed sperm cells were treated with 10 mM dithiothreitol (DTT) along with 0.5% (w/v) SDS in sperm lysis buffer at 37°C for 3 h. Following sperm cell lysis, nuclear preparation was done using Magna ChIP (Magna ChIP A/G immunoprecipitation kit, Cat no. 17–610). Next, the chromatin was sheared using 100 gel units of micrococcal nuclease (NEB, Cat no. MO247S) to get mono or di-nucleosomes. A fraction (∼5%) of sheared chromatin preparations was preserved as whole cell extract (WCE) and the remaining part was immunoprecipitated (IP) using antibody against H3K4me2 and H3K27me3 obtained from Millipore (Cat no. 17–677 and 17–622) as per the Magna ChIP protocol. Following immunoprecipitation, protein/DNA complexes were reverse cross linked and purified. Purity of Immunoprecipitated DNA (IP) and WCE was assessed by the NanoDrop ND-1000 UV–Vis Spectrophotometer (NanoDrop technologies, Rockland). The fragmentation pattern of the WCE DNA was assessed by Agilent Bioanalyzer 2100. A fragmentation range of 200–800 bp was considered as optimal.
| Gene name | Forward sequence | Reverse sequence | Product length | Tm(°) |
|---|---|---|---|---|
| PRM1 | 5′AGATGTCGCAGACGAAGGAG3′ | 5′AGTGCGGTGGTCTTGCTACT3′ | 119 | 60 |
| CD4 | 5′GAGCCTGACCTTGACTTTGG3′ | 5′TGACAGGCTCTTGACGTCTC3′ | 100 | 60 |
| CD45 | 5′AGCCCATTC ATGCAGATGTT3′ | 5′CTGAATACCCGTGGAATGCT3 | 103 | 60 |
| GAPDH | 5′GCCGTAACTTCTGTGCTGTG3′ | 5′AATTAAAAGCAGCCCTGGTG3′ | 109 | 59 |
| PRDM14 | 5′TTCAGGGTAAAGTGGTCAAC 3′ | 5′CACCTCCTTTTCCATCAATA3′ | 114 | 55 |
| SOX4 | 5′CAAAGATAGCGACAAGATCC 3′ | 5′CTTGTAGTCGGGGTAGTCAG3′ | 82 | 55 |
| TBX15 | 5′GATGAGACAGGTGGTCAGTT 3′ | 5′TACTCGAGGCTGGTATTTGT3′ | 106 | 54.9 |
| C-MYC | 5′GAGAAACCTAATAGTGCTTTGG 3′ | 5′GAAGCAGCTCTATTTCTGGA3′ | 82 | 55 |
| PRM2 | 5′CGCTACCACTACAGACACAG 3′ | 5′TCCTCCTCCTCATCCTTC3′ | 115 | 55 |
LIGATION MEDIATED AMPLIFICATION OF WCE AND IP DNA
Agilent ChIP-on-Chip protocol version 10.2 was used to perform this experiment. WCE (250 ng) and IP DNA were subjected to blunting by T4 DNA polymerase at 12°C for 20 min. Post blunting reaction, samples were purified using Phenol/Chloroform/isoamyl alcohol followed by ethanol precipitation. End repaired IP and WCE DNA were ligated with linkers at 16°C for 16 h (Oligos used for preparation of linkers were: 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; 5′-GAATTCAGATC-3′). The ligated DNA samples were precipitated using ethanol suspended in 25 μl of nuclease free water. The linker ligated IP and WCE samples were subjected to two rounds of amplification, LMPCR 1 (15 cycles) and LMPCR 2 (24 cycles). PCR amplified DNA samples were cleaned by salt precipitation and suspended in 25 μl of nuclease free water.
LABELING OF DNA AND HYBRIDIZATION TO MICROARRAYS
Two micrograms each of IP and WCE DNA were labeled using Agilent genomic DNA labeling kit with Cy3 and Cy5 dUTP (5190–0453), respectively. DNA yield and incorporation of labeled dNTPs (specific activity) was measured using NanoDrop spectrophotometer. 3.5 μg of both Cy3 labeled WCE DNA and Cy5 labeled IP samples were hybridized on the custom designed buffalo specific 4 × 180 K ChIP-on-Chip array (Genotypic, Bangalore, India AMADID: 040096). Hybridizations were done using the Agilent hybridization kit (part number: 5188–5308) in Agilent's Sure Hybchambers at 65°C for 24 h. The hybridized slides were washed using Agilent aCGH wash buffers (Part No: 5188–5226) and scanned using Agilent microarray scanner G2505C at three micron resolution.
FEATURE EXTRACTION AND MICROARRAY DATA ANALYSIS
Data extraction from images was done using Feature Extraction Software version 10.7 of Agilent. Feature extracted raw data was analyzed using Agilent Genomic Workbench Lite Edition 6.5 (ChIP-on-Chip application). Normalization of the data was done using Lowess (locally weighted scatter-plot smoothing) normalization method [Zahurak et al., 2007]. Significant genes and regions enriched were identified using Peak Detection Whitehead Per-Array Neighborhood model algorithm by Agilent ChIP analytics software. This method uses the distribution of all probes on each array to compute robust regions of increased probe signal (termed “peaks”). The model samples every probe and its immediate upstream or downstream neighboring probes to identify a robust estimate of the location of bound protein significantly enriched. Derived P values were used to calculate P (Xbar) value, the average P value for the central probe and its neighbors for enriched gene. The probes for which the p(xbar) was more than or equal to 0.055 were identified as enriched for respective histone modifications. Fold values were calculated as the log base two ratio of normalized IP signal to normalized WCE signal for respective genes [Srivastava et al., 2013]. Function and pathway analysis of enriched genes was analyzed using PANTHER classification system (http://www.pantherdb.org). The ChIP-on-Chip array data reported here have been deposited to NCBI's Gene Expression Omnibus [GEO Accession number: GSE49316].
VALIDATION OF ChIP-on-chip DATA BY qPCR EXPERIMENT
For validation purpose the total RNA from sperm samples of each of the bulls (five HF and five SF) were extracted with TRIzol RNA isolation reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol with minor modifications. Briefly, the cryopreserved semen samples (6–7 × 107 sperm) were thawed, washed twice in 12 ml of PBS at 4000 rpm for 5 min, then re-suspended in a somatic cell lysis buffer containing 0.5% (v/v) Triton X-100 and 0.1% (w/v) SDS [Ostermeier et al., 2002; Goodrich et al., 2007] for 10 min at 4°C. The TRIzol reagent was heated at 65°C and the pelleted samples after washing were incubated in it for 30 min at 65°C to completely disintegrate the membranes. Subsequently, the solution was passed three times through a 30 G needle and vortexed vigorously for 30 s and 200 µl of chloroform was added to it. The samples were homogenized by vortexing for 30 s and incubated for 5 min at room temperature followed by a centrifugation at 12,000 g for 15 min at 4°C. The upper aqueous phase was collected and RNA was precipitated using 70% (v/v) ethanol. Finally, RNA samples were dissolved in 10 µl DEPC treated water and a heating step for 15 min at 58°C was performed. For cDNA synthesis 100 ng of total RNA was converted to cDNA using Revert Aid First strand cDNA synthesis kit (Fermentas, Cat#K1621) following manufacturers’ protocol. For transcript abundance analysis in mature sperm, five genes viz. PRDM14, SOX4, TBX15, C-MYC, and PRM2 were selected for qPCR experiment based on their differential histone modification status and involvement in sperm function. GAPDH amplification was used for normalization of qPCR data. The primer sequences have been given in Table II. qPCR was carried out on Mx3005P (Stratagene, Santa Clara, CA) using SYBR green master mix (Maxima, Fermentas, UAB, Lithuania). A universal thermal condition was used for amplification of all genes, that is, initial denaturing of 10 min at 95°C, followed by 45 PCR cycles (denaturing at 95°C for 30 s, annealing at 52–53°C for 30 s, and extension at 72°C for 30 s).
STATISTICAL ANALYSIS
Differences among bulls for percentage of embryonic stages and gene expression were analyzed using analysis of variance (ANOVA) and expressed as mean ± SEM. Statistical tests were deemed significant at P < 0.05. All statistical procedures related to signal peak detection in microarray hybridization are described in the material method section of microarray data analysis.
RESULT
IVF RESULTS FOR BULLS UNDER STUDY
The cryopreserved semen samples from all ten bulls in this study (selected based on conception rate) were subjected to IVF to cross check their differential fertility status. Results of the IVF are presented in Table III. Based on their consistency in cleavage and blastocyst development rates, the sperm samples from three bulls each of HF and LF groups viz. HF2, HF3, and HF5 in HF group and bulls SF1, SF4, and SF5 in SF group were finally selected for ChIP-on-Chip experiments.
| Group | Bull # | # oocytes | Cleavage % | Blastocyst % |
|---|---|---|---|---|
| High Fertile | HF1 | 218 | 69.53(± 0.94)a | 21.50(± 0.35)b |
| HF2 | 211 | 62.00(± 4.24)a | 37.50(± 1.06)a | |
| HF3 | 179 | 66.67(± 1.95)a | 36.33(± 0.51)a | |
| HF4 | 167 | 65.20(± 0.90)a | 24.40(± 1.20)b | |
| HF5 | 152 | 66.20(± 1.00)a | 33.32(± 2.02)a | |
| Sub- Fertile | SF1 | 164 | 56.54(± 0.66)b | 16.90(± 0.90)c |
| SF2 | 213 | 59.00(± 1.67 b | 20.00(± 0.87)d | |
| SF3 | 153 | 63.16(± 1.16)a | 21.17(± 0.97)d | |
| SF4 | 204 | 54.05(± 0.95)b | 18.00(± 1.67)c | |
| SF5 | 197 | 53.55(± 1.65)b | 18.15(± 1.34)c |
- Bull numbers HF2/3/5 and SF1/4/5 were selected for ChIP experiments. Different superscript in cleavage% and blastocyst% columns indicate significant (P < 0.05) difference within the column. HF, High Fertile; SF, Sub-Fertile. IVF experiments for each bull have been repeated four times.
IDENTIFICATION OF DIFFERENTIALLY HISTONE MODIFIED GENES IN HIGH AND SUB FERTILE BUFFALO BULLS
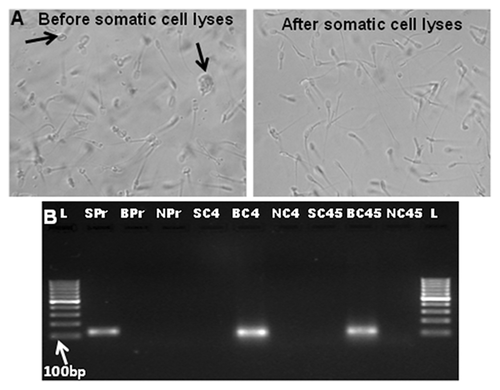
Supporting Table I describe the list of differentially enriched genes for both H3K4me2 and H3K27me3 modified histones in sperm DNA samples of buffalo bulls belonging to the HF and SF groups. Figure 2 depicts the hybridization maps from ChIP-on-chip experiments for respective bulls. Significant genes enriched were identified using peak detection algorithm (event detection) as described in materials and methods. All the probes belonging to a single gene, based on its annotation, was used to calculate their enrichment level for respective histone modifications and then those genes were identified having significant enrichment difference in HF and SF bulls. For selection of differentially enriched genes between the two groups, the genes having enrichment level consistent for all the bulls in respective groups of HF and SF were filtered. Based on this, 84 unique genes for H3K4me2 and 80 for H3K27me3 were identified which exhibited a significant (P < 0.01) difference between HF and SF groups. The identified genes were annotated with reference to bovine genome and then classified on the basis of gene ontology classification.
FUNCTIONAL ANALYSIS OF DIFFERENTIALLY ENRICHED GENES
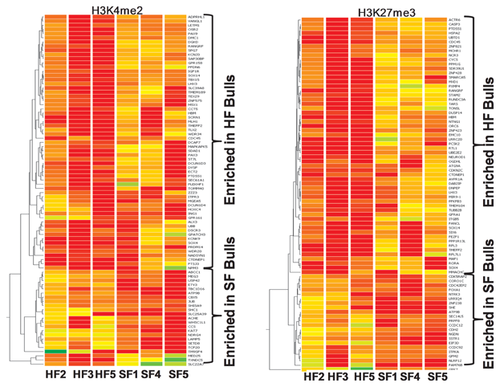
The functional annotations of differentially enriched genes (84 genes for H3K4me2 and 80 genes for H3K27me3) are shown in Figure 3. As per their gene ontology classification the identified genes were classified under three categories viz. biological process, molecular function and cellular component. Functional analysis revealed that by far majority of the differentially enriched genes were involved in molecular binding (25–32%), cell communication (6–25%), cellular process (12–14%), and catalytic activities (7–18%) for both H3K4me2 and H3K27me3 kind of modifications. In biological process group majority of the differentially H3K4me2 enriched genes were involved in metabolic process (25%), transport (14%) and development process (13%) while H3K27me3 enriched genes were mostly involved in cell communication (25%) and cellular process (14%). The most important molecular functions influenced by differential H3K4me2 enrichment were molecular binding (32%), transcription regulation (20%) and catalysis (18%) while in H3K27me3 group most important molecular functions were translation (33%), binding (25%) and receptor (16%). In cellular components group, most of the genes found enriched with H3K4me2 modification belonged to intracellular region (80%) while in H3K27me3 group most important component influenced with differential enrichment was protein complex (57%). Other important functions in which the differentially enriched genes were found involved were reproduction, receptor activity, enzyme regulation, and apoptosis.
DECIPHERING GENES RELATED TO SPERM FUNCTION AND EMBRYONIC DEVELOPMENT
The differentially enriched genes in both H3K4me2 and H3K27me3 experiments were further searched for their potential involvement in sperm function and embryogenesis. These differentially enriched genes in both the lists were found having crucial roles in germ cell development, fertilization and embryogenesis. The genes identified in the H3K4me2 list viz. CDC45, CCT5, DMC1, MLH1, PAX3, PRDM14, SOX4, SOX14, TBX15, and MEG3 were found to have important roles in sperm cell development, different functional attributes of sperm and embryogenesis (Table IV). Also in H3K27me3 group CDKN2C, FANCL, GFRA1, LHX3, RPL3, SIX6, SOX14, SOX4, and FOXA1 genes were found to be involved in sperm function and embryonic development (Table V).
| Gene | Description | P[Xbar] value | Fertility group | Function |
|---|---|---|---|---|
| CDC45 | Cell division cycle 45 | 0.0011 | Enriched in HF bulls | Required for initiation of chromosomal DNA replication [Owens et al., 1997] |
| DMC1 | Disrupted Meiotic CDNA1 | 0.0092 | Participate in meiotic recombination | |
| MLH1 | mutL homolog 1 | 0.0095 | MLH1 mutations as source of human infertility [Hoffmann et al., 2003] | |
| CCT5 | Chaperonin containing TCP1, subunit 5 | 0.0168 | Play a role in the folding of actin and tubulin [Soue‘s et al., 2003] | |
| PAX3 | Paired box 3 | 0.0103 | Transcription factors, plays essential roles during embryogenesis [Underhill, 2000] | |
| PRDM14 | PR domain containing 14 | 0.0028 | Possess histone methyltransferase activity. Play an essential role in germ cell development [Yamaji et al., 2008; Ying et al., 2008] | |
| SOX4 | SRY (sex determining region Y)-box 4) | 0.0028 | Transcription factors involved in the regulation of embryonic development and in the determination of the cell fate [Kamachi et al., 2000] | |
| SOX14 | SRY (sex determining region Y)-box 14 | 0.0180 | Intronless gene, encodes a member of transcription factors, involved in the regulation of embryonic development and in the determination of the cell fate [Kamachi et al., 2000] | |
| TBX15 | T-box protein 15 | 0.0187 | Regulate a variety of developmental processes [Naiche et al., 2005] | |
| MEG3 | Maternally expressed 3 | 0.0073 | Enriched in SF bulls | Maternally expressed imprinted gene, abnormal spermatogenesis is associated with defective MEG3 methylation [Hammoud et al., 2009] |
| Gene | Description | P[Xbar] value | Fertility group | Function |
|---|---|---|---|---|
| CDKN2C | Cyclin-dependent kinase inhibitor 2 C (p18, inhibits CDK4) | 0.0061 | Enriched in HF bulls | Regulation of spermatogenesis [Zindy et al., 2001] |
| FANCL | Fanconi anemia, complementation group L | 0.0233 | Required for proper primordial germ cell proliferation in the embryonic stage [Cheng et al., 2000] | |
| GFRA1 | GDNF family receptor alpha 1 | 0.0034 | Silencing in mouse spermatogonial stem cells results in their differentiation [He et al., 2007] | |
| LHX3 | LIM homeobox 3 | 0.0154 | Mutations causes growth, fertility, and metabolic problems [Mullen et al., 2012] | |
| RPL3 | Ribosomal protein L3 | 0.0030 | Required for embryonic and larval viability, fertility [Mitrovich et al., 2000] | |
| SIX6 | SIX homeobox 6 | 0.0119 | Regulation of GnRH expression and hypothalamic control of fertility [Larder et al., 2011] | |
| SOX14 | SRY (sex determining region Y)-box 14 | 0.0078 | Involved in the regulation of embryonic development and in the determination of the cell fate [Kamachi et al., 2000] | |
| SOX4 | SRY (sex determining region Y)-box 4 | 0.0007 | Involved in the regulation of embryonic development and in the determination of the cell fate [Kamachi et al., 2000] | |
| FOXA1 | Forkhead box A1 | 0.0119 | Enriched in SF bulls | Transcription factor involved in development process, establishment of tissue-specific gene expression [Lupien et al., 2008] |
TRANSCRIPT ABUNDANCE OF SELECTED GENES
Interpretations from the ChIP-on-Chip experiments were validated by amplification of transcripts of five genes found differentially modified in HF vs. SF bulls and having role in sperm function and embryonic development. The presence of PRDM14, SOX4, and C-MYC transcripts were found to be present in mature sperm while the transcripts of TBX15 and PRM2 were not detectable (Fig. 4). There was a significant (P < 0.05) difference in the abundance of SOX4 in HF vs. SF bulls.

DISCUSSION
The differential physiologic measurements of sperm function like morphology, motility, membrane integrity, etc. often do not explain the underlying cause of male infertility [Moldenhauer et al., 2003]. The current study was undertaken to explore the whole genome histone modification differences between HF and SF buffalo bulls with the expectation that it would generate some clues about the molecular reasons for differential sperm fertilizing ability and hence the bull fertility. In view of the fact that the spermatogenesis events goes through a very dynamic chromatin modulation process [Carrell and Hammoud, 2010] we considered that the ChIP-on-chip assessment of mature sperm for the selected histone modifications may provide a meaningful reflection of the fidelity of the past events during spermatogenesis, eventually explaining the overall fertility status of bulls. The study focused on two kinds of histone modifications viz. H3K4me2 and H3K27me3 as they represent the activating and repressing kind of histone modifications, respectively. Earlier evidences support that human sperm show strong enrichments of both of these kinds of modified histones at regulatory regions of genes related to embryonic development [Hammoud et al., 2009; Brykczynska et al., 2010]. H3K4me2 has been found enriched at the promoters of developmentally important genes, whereas tri-methylated H3K27 (H3K27me3) is significantly enriched at promoters that are repressed in early embryos but get expressed during blastocyst development and implantation, including many bivalent (H3K4me3/H3K27me3) promoters which are important for embryonic stem cell development [Hammoud et al., 2009].
The microarray data analysis revealed 84 genes in H3K4me2 and 80 genes for H3K27me3 group as differentially enriched in sperm from HF and SF buffalo bulls (Supporting Table S1). Out of these genes ten genes in H3K4me2 and nine genes in H3K27me3 lists have been found to be involved in sperm function and embryogenesis (Table IV & V). Table IV include cell cycle regulatory genes like CDC45, DMC1, and MLH1 which have been reported to be involved in chromosomal DNA replication, meiotic recombination etc. [Owens et al., 1997; Hoffmann et al., 2003]. The MLH1 has at least three meiotic functions viz. heteroduplex repair, crossing over and chromosome segregation and mutations in this gene have been associated with human infertility [Hoffmann et al., 2003]. In our ChIP-on-Chip experiment, all these genes were found to be H3K4me2 enriched in sperm of HF bulls. Considering our results and the fact that H3K4me2 histone modifications impact transcriptional activation of the associated genes, the possible role of CDC45, DMC1, and MLH1 can be implicated for imparting better fertilizing ability to the sperm of HF bulls. The role of CCT5 in folding of tubulin and actin proteins, required for sperm tail development has been described [Soue‘s et al., 2003]. We have detected this gene to be H3K4me2 enriched in HF bulls. Since H3K4me2 modification represents a histone activation process and CCT5 is reportedly involved in sperm motility, our observations further consolidates its potential role in imparting better motility to the sperm of HF bulls. Similarly, we observed MEG3 lacking H3K4me2 enrichment in HF bulls. A defective MEG3 (also known as GTL2) has been associated with abnormal spermatogenesis [Hammoud et al., 2009]. The spermatozoa have been reported to carry both activating and repressing histones with bivalent modification [Takada et al., 2000; da Rocha et al., 2008; Glazov et al., 2008]. Notably, the promoter of MEG3 lacks DNA methylation in sperm, but re-acquires it during embryogenesis. Also, it is a maternally expressed gene. In our experiment this gene was found lacking in H3K4me2 enrichment in HF bulls which favors the logic of its transcriptional inactivity in HF bulls for normal spermatogenesis. The third set of genes found H3K4me2 enriched in HF bulls consisted of the genes involved in embryonic and fetal development viz. PAX3, PRDM14, SOX4, SOX14, and TBX15. PAX3 is a member of the paired box (PAX) family of transcription factors which plays essential role during embryogenesis [Underhill, 2000]. Mutation-deletion studies in Drosophila indicate that the N-terminal region of PAX3 is involved in determining male fertility [Xue et al., 2001]. PRDM14 has been reported to possess histone methyltransferase activity and is required for the maintenance of embryonic stem cell identity and the reacquisition of pluripotency in somatic cells [Yamaji et al., 2008]. They have also been shown to play essential role in germ cell development [Ying et al., 2008]. SOX4 and SOX14 are members of the SRY-related HMG-box family of transcription factors, involved in the regulation of embryonic development and in the determination of the cell lineage [Kamachi et al., 2000]. TBX15 is required in a diverse array of developmental processes in post-implantation embryo including formation and patterning of the mesoderm and organogenesis [Naiche et al., 2005].The histone modification status of these genes observed in HF bulls of present study (Table IV) hint them as possible candidates to be explored for explaining the fertility status of bulls.
The ChIP-on-Chip data for H3K27me3 also identified important genes related to spermatogenesis, fertility and embryonic development (Table V). We observed that the gene CDKN2C significantly more enriched for H3K27me3 in the sperm of HF bulls. Earlier knockout studies in mice have indicated that, for effective regulation of spermatogenesis, this gene has to remain suppressed to ensure the mitotic exit and the normal maturation of spermatocytes [Zindy et al., 2001]. Likewise, FANCL and GFRA1 were also found H3K27me3 enriched in mature sperm of HF bulls supporting their role in primordial germ cell proliferation and differentiation during early phase of spermatogenesis [Cheng et al., 2000; He et al., 2007].
Retained histones and their modified forms in sperm genome have been shown to be significantly enriched at promoters of developmentally important genes [Hammoud et al., 2009]. Consolidating their role in embryonic development our results revealed higher enrichment for both H3K4me2 and H3K27me3 modifications of SOX4 and SOX14 in sperm of HF bulls [Kamachi et al., 2000]. FOXA1, found enriched with H3K27me3 in SF bulls in the present experiment, is a transcription factor involved in embryonic development and establishment of tissue-specific gene expression [Lupien et al., 2008]. Supporting our results earlier reports have also evidenced the role of LHX3, RPL3, and SIX6 in fertility through pituitary and hypothalamic pathways [Mitrovich and Anderson, 2000; Larder et al., 2011; Mullen et al., 2012].
Taking leads from our ChIP-on-chip results and considering the role of differentially modified genes in sperm function, the PRDM14, C-MYC, TBX-15, PRM2, and SOX4 genes were selected to verify whether the histone modifications result in their differential transcript abundance in mature sperm of HF and SF buffalo bulls. Results are explained in Figure 4. While we could detect the presence of PRDM14, C-MYC, and SOX4 transcripts in mature sperm from all the bulls under study, the PRM2 and TBX15 transcripts were undetectable. The significant (P < 0.05) difference of SOX4 transcript abundance observed in HF vs. SF bulls was in support of its role in embryonic development [Kamachi et al., 2000]. However, considering the fact that the mature sperm as such represents a population of transcriptionally inert cells [Kierszenbaum and Tres, 1975], conclusive interpretation out of this set of experiment is unwarranted.
The findings of the current study suggest that differential methylation rates of retained histones in the mature sperm of buffalo bulls could be of potential significance for their fertility performance. The growing field of sperm epigenome carriage vis-à-vis fertilizing ability of spermatozoa will benefit our understanding of the fertility problems of breeding bulls. Considering the importance of farm animal species as model organisms and similarities of their reproduction physiology, the knowledge generated in the present work will possibly benefit investigations in the context of human infertility also. To our knowledge, this is the first attempt to employ the high throughput microarray technology for studying the epigenomic status of buffalo sperm in relation to bull fertility.
ACKNOWLEDGMENT
The financial aid under NAE project of ICAR to T.K.D. is thankfully acknowledged.