Long interspersed nucleotide element-1 hypomethylation in folate-deficient mouse embryonic stem cells†
Shaoyan Chang and Li Wang contributed equally to this work.
Abstract
Folate is thought to contribute to health and development by methylation regulation. Long interspersed nucleotide element-1 (LINE-1), which is regulated by methylation modification, plays an important role in sculpting the structure and function of genomes. Some studies have shown that folate concentration is related to LINE-1 methylation. However, the direct association between LINE-1 methylation and folate deficiency remains unclear. To explore whether folate deficiency directly induced LINE-1 hypomethylation and to analyze the relationship between folate concentration and the LINE-1 methylation level, mouse ESCs were treated with various concentrations of folate which was measured by chemiluminescent immunoassay, and the homocysteine content was detected by ELISA. LINE-1 methylation was examined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry at various time points. Concurrently, cell proliferation and differentiation were observed. The result showed that the intracellular folate decreases under folate-deficient condition, conversely, homocysteine content increased gradually and there was a negatively correlated between them. Folate insufficiency induced LINE-1 hypomethylation at the lowest levels in folate-free group and moderate in folate-deficient group, compared with that in the folate-normal group at day 18. Moreover, LINE-1 methylation level was positively correlated with folate content, and negatively correlated with homocysteine content. At corresponding time points, proliferation and differentiation of mouse ESCs showed no alteration in all groups. Our data indicated that folate deficiency affected the homeostasis of folate-mediated one-carbon metabolism, leading to reduced LINE-1 methylation in mouse ESCs. This study provides preliminary evidence of folate deficiency affecting early embryonic development. J. Cell. Biochem. 114: 1549–1558, 2013. © 2013 Wiley Periodicals, Inc.
Folate is involved in DNA synthesis and methylation as a mediator for transfer of one-carbon units, and plays crucial roles in normal cell function, embryonic development and tumorigenesis [Fox and Stover, 2008]. Former studies have shown that one-carbon units metabolic disorders caused by folate deficiency can affect the genome methylation level, and eventually result in abnormal development and diseases [Crider et al., 2012]. An epidemiological study has shown that a folate-deficient diet in older women is associated with decreased genomic DNA methylation in leukocytes [Rampersaud et al., 2000]. Similar results have also been found in postmenopausal women with a folate-deficient diet, in which increased homocysteine concentrations and decreased genomic methylation in lymphocytes and the global methylation level were reversed during a 3-week period of folate supplementation [Jacob et al., 1998]. Some studies of cancer have also found that genomic methylation in the colonic mucosa is increased in response to folate supplements taken by colorectal adenoma and colon cancer patients [Cravo et al., 1994, 1998; Kim et al., 2001]. Moreover, an animal model study has shown that a folate-free diet in mice leads to reduced liver genomic methylation [Pogribny et al., 1995]. Therefore, folate influences genomic methylation, although whether folate deficiency has a direct effect on methylation modification of specific genes or regions is unclear.
Long interspersed nucleotide element-1 (LINE-1) is abundant in the genome at about 20% [Kazazian, 2004] and has an autonomous transposition activity. Some studies have demonstrated that a low level of folate is related to LINE-1 methylation disorders. In low birth weights, there is a positive correlation between folate deficiency and LINE-1 hypomethylation in cord blood [Fryer et al., 2011]. Some cancer studies have also found that folate deficiency results in lower LINE-1 methylation. Jin et al. [2009] found that folate levels are lower in lung cancer patients, meanwhile LINE-1 methylation levels are significantly lower than those in normal individuals, a positive correlation exists between folate and LINE-1 methylation, and folate supplements can prevent the occurrence of lung cancer.
LINE-1 transposition activity is regulated by DNA methylation in which an elevated degree of LINE-1 methylation causes inactive transposition [Iskow et al., 2010]. For example, LINE-1-mediated retrotransposition events can act as mutagens [Belancio et al., 2008; Goodier and Kazazian, 2008], affect gene expression [Perepelitsa-Belancio and Deininger, 2003; Han et al., 2004] and are required to silence chromosome X by Xist [Chow et al., 2010]. The transposable role of LINE-1 is active and heritable, particularly in germ cells and embryonic stem cell (ESC) lines. Thus, LINE-1 plays a critical role in the process of embryonic development [Perepelitsa-Belancio and Deininger, 2003; Han et al., 2004; Belancio et al., 2008].
Our previous studies have found lower genome and LINE-1 methylation levels in neural tube defects of embryos, which is accompanied by a reduced folate concentration and elevated homocysteine concentration in maternal plasma. We have showed that genome and LINE-1 methylation levels are related to the risk of neural tube defects, but no correlation between folate and the LINE-1 methylation was observed. To date, whether abnormal folate level affects the methylation of genome and LINE-1 during embryonic development has not been studied. Therefore, we established a cell model of folate deficiency in mouse ESCs that are derived from the inner cell mass of embryos. We examined LINE-1 methylation by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) following sodium bisulfite conversion to analyze whether folate deficiency may be relevant to aberrant LINE-1 methylation, and explored the correlation between folate and LINE-1 methylation during early embryonic development.
MATERIALS AND METHODS
Cell Culture
ESC culture
Mouse ESCs, Sv/129, were obtained from Xuanwu Hospital (Beijing, China), and maintained on mitotically inactivated primary mouse embryonic fibroblasts prior to cultivation under feeder-free conditions. After seeding onto culture dishes coated with 0.2% gelatin (Sigma-Aldrich, MO, USA), mouse ESCs were maintained in folate-free Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 0.1 mM β-mercaptoethanol, non-essential amino acids, 2 mM glutamate, 15% fetal bovine serum (all purchased from Invitrogen, Carlsbad, USA), 4 mg/l folate (Sigma-Aldrich) and 1,000 U/ml leukemia inhibitory factor (Millipore, Billerica, USA). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2, and passaged every 3 days. Medium changes were performed daily.
Folate Treatments of ESC Cultures
After three passages under folate-normal condition, mouse ESCs were cultured in medium with various folate concentrations, and were divided into three groups treated with 4 mg/l folate (folate-normal group, defined as FN), 0.5 mg/l folate (folate-deficient group, defined as FD) or without folate (folate-free group, defined as FF).
Biochemical Analyses
Folate content in Mouse ESCs was measured by using a competitive receptor binding immunoassay (Chemiluminescent Immunoenzyme Assay Access Immunoassay System II; Beckman Coulter, Krefeld, Germany). The homocysteine content was detected using a homocysteine ELISA kit according to the manufacturer's instructions (Dongge, Beijing, China).
LINE-1 Methylation Analysis
DNA extraction
Genomic DNA was extracted from 5 × 106 cells with a Blood and Cell Culture DNA Kit (QIAGEN, Dusseldorf, Germany) according to the manufacturer's instructions. No RNA contamination was detected by agarose gel electrophoresis. Final preparations had an A260:A280 ratio of about 1.8.
Bisulfite treatment
A total of 500 ng genomic DNA from each sample was subjected to bisulfite treatment with a Methylamp DNA Modification Kit (Epigentek, NY, USA). The quality of the bisulfite conversion was controlled using PCR products without methylation. Sequencing results confirmed that 96.6% of cytosine residues were converted.
Methylation measure and analysis
A Sequenom MassARRAY platform (CapitalBio, Beijing, China) was used to perform the quantitative methylation analysis of LINE-1 (GenBank Accession Number: D84391). The robustness of this approach for quantifying methylated and unmethylated DNA has been validated by sequencing in our laboratory [Wang et al., 2010]. This system uses MALDI-TOF mass spectrometry in combination with RNA base-specific cleavage (MassCLEAVE). A detectable pattern is then analyzed for its methylation status. PCR primers used in this study have been described elsewhere [Kuramochi-Miyagawa et al., 2008]. For each reverse primer, an additional T7 promoter tag for in vivo transcription was added, and a 10-mer tag was added onto the forward primer to adjust for melting temperature differences. The following primers were used to amplify nt 874–1,156 of the LINE-1 promoter: 5′-aggaagagag GTTAGAGAATTTGATAGTTTTTGGAATAGG-3′ and 3′-cagtaatacgactcactatagggagaaggct CAAAACAAAACCTTTCTCAAACACTATAT-5′. In total, 10 CpG sites, which were divided into nine CpG units, were examined in this area, except for the third and fourth CpG sites that were indistinguishable (Fig. 2). The product was added into Shrimp alkaline phosphatase (SAP; Sequenom, San Diego, CA) to remove dNTPs. T7 R&DNA polymerase (Epicentre, Madison, WI) were used to incorporate thymidine triphosphate in the transcripts. Meanwhile RNase-A (Sequenom) was added to cleave the in vitro transcripts (T-cleavage assay). The product was added into resin to prepare for MassARRAY. Spectra methylation ratios were generated by Epityper software version 1.0 (Sequenom).
Cell Proliferation and Apoptosis Analyses
Cell proliferation assay and cell cycle analysis
Cells were incubated and processed with a BrdU Labeling and Detection Kit III (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. Cells were stained with BrdU, anti-BrdU-PODS, peroxidase substrate, Cy2-conjugated AffiniPure Sheep Anti-Mouse IgG (Lot #: 515-225-003; Jackson, USA), and a 0.001% 4,6-diamidino-2-phenylindole (DAPI; Lot #: 32670; Fluka, USA) solution for nuclear staining. The proliferative index was calculated as the ratio of BrdU-positive nuclei (green fluorescence) to the total number of nuclei (blue fluorescence) under a fluorescence microscope (Olympus, Tokyo, Japan). Cells from the same population that were not BrdU-labeled were used as a negative control.
Mouse ESCs were rinsed twice with phosphate-buffered saline (PBS) and digested with 0.25% trypsin. After centrifugation for 3 min at 1,000g, the supernatant was removed and cells were resuspended and fixed with 70% cold ethanol. Fixed cells were stained with 50 µg/ml propidium iodide containing 0.1% Triton X-100 and 0.02 mg/ml ethylenediaminetetraacetic acid (EDTA), and then analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). A total of 10,000 events were recorded for each sample.
Stage-specific embryonic antigen-1 (SSEA-1) assay
For immunocytochemistry, cultured cells were fixed with 4% paraformaldehyde in 0.1 M PBS, and then permeabilized for 10 min with 0.2% Triton X-100. PBS containing 10% normal goat serum was used a blocking solution, and then cells were incubated overnight with a primary anti-SSEA-1 antibody (1:50; Santa Cruz Biotechnology, CA, USA) at 4°C. Cells were then incubated for 1 h with a secondary Cy2-conjugated AffiniPure Sheep Anti-Mouse IgG and a 0.001% DAPI solution for nuclear staining. SSEA-1 was observed under a fluorescence microscope and analyzed by flow cytometry.
Pluripotent genes expression
Total RNA was extracted from cultured cells using Trizol reagent (Invitrogen) according to the manufacturer's instructions, and then treated with DNase (Promega, Madison, USA) to remove contaminating DNA. RNA (1 µg) was reverse transcribed into cDNA using random primers and Superscript III™ (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed with a 7500 Fast Real-Time PCR System (Applied Biosystems). The following primers sequences were designed using PrimerExpress software (Applied Biosystems). Oct4, forward 5′-GTGGAAAGCAACTCAGAG-3′, and reverse 5′-CTTGGCAAACTGTTCTAGC-3′; Nanog, forward 5′-TGCTACTGAGATGCTCTG-3′ and reverse 5′-TTGTTCTCCTCCTCCTCA-3′; Sox2, forward 5′-GAAACTTTTGTCCGAGACC-3′ and reverse 5′-GCGTGTACTTATCCTTCTTC-3′; and β-actin, forward 5′-CATTGTTACCAACTGGGAC-3′ and reverse 5′-GATCTGGGTCATCTTTTCAC-3′. PCR cycling parameters were 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. Samples for PCR were run in triplicate. All samples were normalized against β-actin using the comparative CT method.
Statistical Analyses
Data were analyzed with the SPSS 16.0 software package (McGraw-Hill, Inc., New York, NY). One-way analysis of variance was performed to evaluate the significance of any differences among FF, FD, and FN. Correlation analysis was performed using bivariate correlations. Pearson's correlation coefficient (r) was used to evaluate the correlation between folate concentration and LINE-1 methylation, and LINE-1 methylation and homocysteine concentration to reflect the degree to correlations. Data were presented as the mean and standard deviation (SD) and significance was accepted at P < 0.05.
RESULTS
Folate Content in Mouse ESCs and Effect of Folate Deficiency on Homocysteine Concentration
Mouse ESCs were cultured in medium with various folate concentrations and intracellular folate content was measured by Chemiluminescence method. The results showed that folate defection in medium have caused decreased intracellular folate concentration in 6th days of treatment, as shown in Figure 1, folate content was 0.32 ng/106 cells in FF, which was less than in FD (0.96 ng/106 cells), or that in FN (2.08 ng/106 cells). Significant differences still existed among groups over time, although no more decreased folate concentration were detected in any group of folate defection. Homocysteine is a key intermediate metabolite of folate metabolism, therefore, we measured the homocysteine concentration of mouse ESCs cultured in medium with various folate concentrations by ELISA. As shown in Figures 1-3, the homocysteine content increased obviously in folate-deficient groups over time, and the FF was higher than the FD. The value was 0.31 nmol/1 × 105 cells at day 6 increasing to 0.36 nmol/1 × 105 cells at day 18 in FF, 0.29 nmol/1 × 105 to 0.33 nmol/1 × 105 cells in FD, but no alteration was observed in FN, as 0.28 nmol/1 × 105 cells. Further analysis of the results showed a significant negative correlation between folate and homocysteine content.
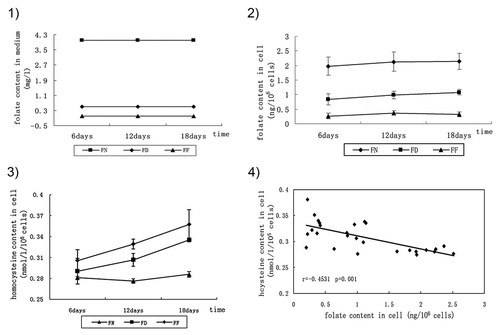
Folate and homocysteine content at various times points and the relevance of them. (1) Folate concentration in medium. (2) Folate content of mouse ESCs cultured in medium with various folate concentrations, as measured by a competitive receptor binding immunoassay. (3) Homocysteine content of mouse ESCs cultured in medium with various folate concentrations, as measured by ELISA. (4) Correlation of folate content with homocysteine content in cell. Bivariate correlation was performed. CpG sites were numbered 1–10 from the 5′ to 3′ end in the 5′UTR of LINE-1. Pearson's correlation coefficient was performed. Folate treatment was categorized as FN, FD, and FF. FF was used as a control. FF, folate-free group; FD, folate-deficient group; FN, folate-normal group.
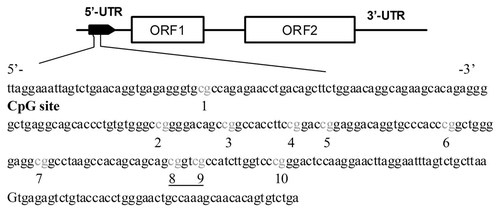
Schematic diagram of LINE-1. LINE-1 transcript consists of a 5′-untranslated region (5′-UTR) with internal promoter activity, two open reading frames (ORF1 and ORF2), and a 3′-UTR. The sequence shown represents a 283-bp fragment (nt 874–1,156) in the 5′-UTR of LINE-1. Numbers 1–10 indicate CpG sites within the LINE-1 elements tested, and underlines highlight CpG units that include more than one CpG site tested at the same time.
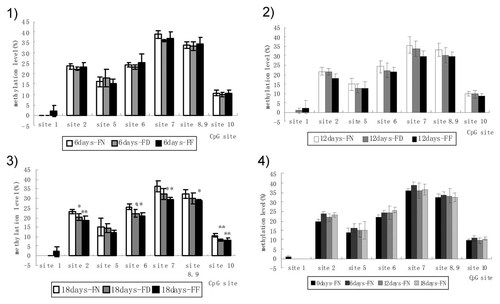
Methylation levels of LINE-1 among groups at various time points, as detected by mass spectrometry. Methylation levels of all CpG sites in LINE-1 were compared among FN, FD, and FF. (1)–(3) LINE-1 methylation level among groups at 6, 12, and 18 days. (4) LINE-1 methylation level in the control group at 0, 6, 12, and 18 days. CpG sites were numbered 1–10 from the 5′ to 3′ end in the 5′ of LINE-1. FF, folate-free group; FD, folate-deficient group; FN, folate-normal group. Data are the mean and SD. *P < 0.05, **P < 0.01 (relative to the respective control).
Effect of Folate Deficiency on LINE-1 Methylation
To determine whether DNA methylation was affected by folate, we measured LINE-1 methylation in mouse ESCs by MALDI-TOF of various groups treated with folate. Mouse ESCs were cultured in folate-free medium, folate-deficient medium, and folate-normal medium for 6, 12, and 18 days. As shown in Figure 2, the LINE-1 methylation level showed little difference among the three groups at day 6, but an emerging trend of low LINE-1 methylation was found in FF and FD at days 12 and 18, compared with that in FN, which was more evident in FF than in FD. Especially at days 18, most CpG sites of LINE-1 showed significant differences in FF and FD compared with that in FN, which was more evident in FF than in FD (P < 0.05 or P < 0.01).
Correlation of Folate and Homocysteine Content With LINE-1 Methylation Levels, Respectively
To establish the relationship between LINE-1 methylation and intracellular folate content or homocysteine content, Pearson's correlation was used coefficient to determine the significant correlation, respectively. The result showed that folate content was positively related to LINE-1 methylation (r = 0.481, P < 0.05, Figs. 4-7) and homocysteine content was negatively related to LINE-1 methylation in mouse ESCs (r = –0.54, P < 0.01, Figs. 5-7). Further analysis revealed that each CpG site of LINE-1 had a significantly positive correlation between folate content and LINE-1 methylation level, except for the fifth and the eighth–ninth CpG sites (Fig. 4) and negative correlation between homocysteine and LINE-1 methylation levels, except for the fifth CpG site (Fig. 5).
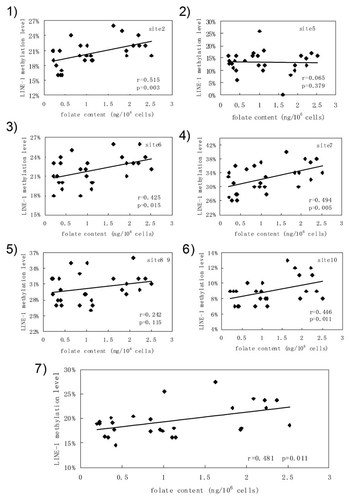
The relevance between LINE-1 methylation and folate content. (1)–(6) Correlation of CpG methylation levels in LINE-1 and folate content. (7) Correlation of the LINE-1 methylation level with folate content. CpG sites were numbered 1–10 from the 5′ to 3′ end in the 5′UTR of LINE-1. Pearson's correlation coefficient was performed.
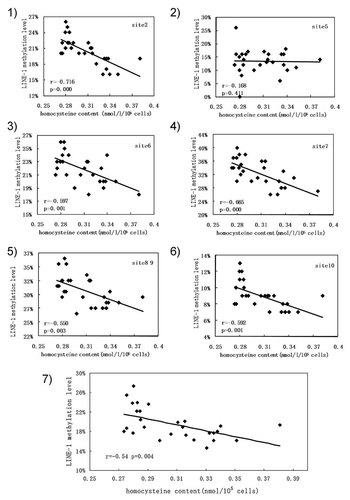
The relevance between LINE-1 methylation and homocysteine content. (1)–(6) Correlation of CpG methylation levels in LINE-1 and homocysteine content. (7) Correlation of the LINE-1 methylation level with homocysteine content. CpG sites were numbered 1–10 from the 5′ to 3′ end in the 5′UTR of LINE-1. Pearson's correlation coefficient was performed.
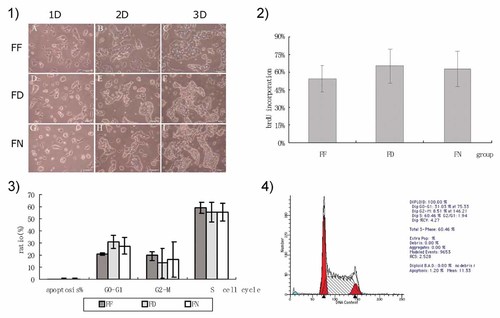
Proliferation and apoptosis of mouse ESCs in medium containing various folate concentrations. (1) Cell morphology as indicated by microscopy. (2) Cell proliferation as indicated by BrdU incorporation. (3) Cell cycle analysis by flow cytometry. (4) Histogram of flow cytometry data. FF, folate-free group; FD, folate-deficient group; FN, folate-normal group; 1, 2, and 3 days: culture days of mouse ESCs.
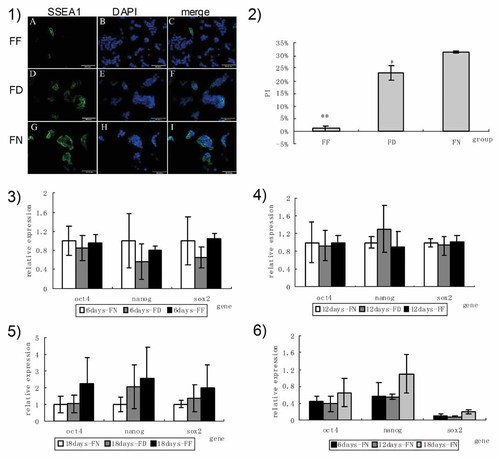
Expression of SSEA1 and pluripotency genes in mouse ESCs cultured in medium with various folate concentrations. We detected the expression level of SSEA-1 in mouse ESCs cultured in folate-free, -deficient, and -normal mediums by immunofluorescent staining (1) and flow cytometric analysis (2). Pluripotency gene expression levels of Oct4, Nanog, and Sox2 in mouse ESCs were compared among FN, FD, and FF by real-time PCR. (3)–(5) Expression levels of Oct4, Nanog, and Sox2 in mouse ESCs in FN, FD, and FF at 6, 12, and 18 days. (6) Expression level of Oct4, Nanog, and Sox2 in the control group at 6, 12, and 18 days. FF, folate-free group; FD, folate-deficient group; FN, folate-normal group; SSEA-1, stage-specific embryonic antigen-1; DAPI, 4,6-diamidino-2-phenylindole, nucleus staining.
Effect of Folate Deficiency on Mouse ESC Proliferation and Differentiation
Because folate participates in DNA synthesis, to analyze whether folate deficiency had an effect on mouse ESC proliferation, we evaluated mouse ESC morphology by microscopy, proliferation by BrdU incorporation, and apoptosis by flow cytometry. The results showed no significant difference in the cell proliferation of FF and FD, compared with that of FN (Fig. 6).
SSEA-1, a marker of mouse ESCs, showed different expression levels in mouse ESCs cultured in folate-free and -deficient medium, compared with that in folate-normal medium. As shown in Figure 7, SSEA-1 expression levels were the lowest in FF, intermediate in mouse ESCs cultured in FD, compared with those in FN.
The effect of folate deficiency on mouse ESC differentiation was determined by real-time PCR. Oct4, Sox2, and Nanog are key components of the pluripotency network of ESCs, which regulate mouse ESC self-renewal and differentiation. Therefore, we measured their expression levels in mouse ESCs cultured in various medium for 6, 12, and 18 days to evaluate differentiation. As shown in Figure 7, there was no change among groups at days 6 and 12. Until 18 days, an increasing tendency of expression levels of Oct4, Sox2, and Nanog was observed, which was higher in FF than that in FD compared with that in FN, but showed no significant difference among groups.
DISCUSSION
In this study, a significantly negatively correlation was observed between intracellular folate and homocysteine content in mouse ESCs cultured in folate-deficient medium. Meanwhile, we found reduced LINE-1 methylation level in mouse ESCs cultured in folate-deficient medium, and LINE-1 methylation was positively correlated with folate content and negatively correlated with homocysteine content. These results indicate that folate metabolism plays a role in acquiring and/or maintaining LINE-1 methylation in early embryonic development. A low folate concentration probably causes aberrant LINE-1 methylation, thereby affecting early embryonic development.
Mouse ESCs cultured in a low folate condition may possess an abnormal folate metabolism, leading to disorder of intracellular folate metabolites. In our results, decreased intracellular folate content in mouse ESCs cultured with folate-deficient medium was confirmed by ACCESS II assay, homocysteine concentration was also detected simultaneously by ELISA. Homocysteine is an important product of folate metabolism [Kim et al., 2009]. Homocysteine accumulation in cells can be found when folate deficiency [Kim et al., 1997]. In this study, we found that the homocysteine content in mouse ESCs increased gradually accompanied by decreasing of folate concentration over time, and a negative correlation between them was observed. These results suggested that folate deficiency during cell proliferation led to abnormal folate metabolism in cells.
Our results showed that folate deficiency led to reduction of LINE-1 methylation in mouse ESCs with low folate treatment, and this reduction in FF was less than in FD. The LINE-1 methylation level reduced gradually over time. Meanwhile a positive correlation between folate content and LINE-1 methylation level was found. These results indicated that aberrant LINE-1 methylation may be directly effected by folate concentration in mouse ESCs and a definite causal relationship existed between LINE-1 methylation modification and folate metabolism. Jin et al. [2009] found that LINE-1 methylation was significantly reduced in the lung cancer compared with that in the normal tissue, and correlated positively with folate concentration. In colon cancer patients, a low folate diet is positively correlated with LINE-1 methylation [Schernhammer et al., 2009]. These epidemiological findings suggest that folate was positively correlated with LINE-1 methylation in tumor tissue, but whether aberrant LINE-1 methylation is caused directly by folate deficiency is unclear. In this study, we found that LINE-1 methylation significantly decreased in mouse ESCs with folate treatments (Fig. 3), and a positive correlation was observed between intracellular folate content with LINE-1 methylation level (Fig. 4), which suggested that folate may be a direct effect on LINE-1 methylation level of embryonic tissue.
Homocysteine is regarded as an accurate inverse indicator of methylation process which is involved in folate metabolism [Kim, 2004]. Abnormally low circulating levels of folate can cause an accumulation of homocysteine, with a concomitant reduction in S-adenosyl methionine, leading to an impaired and possibly insufficient capacity for DNA methylation [Kim et al., 1997]. Our study found elevated homocysteine content in mouse ESCs cultured in low folate, and a negative correlation between homocysteine content with LINE-1 methylation. Furthermore, the homocysteine concentration and methylation of each CpG site of LINE-1 had a negative relevance (Fig. 5). These results indicate that folate deficiency affects the homeostasis of folate-mediated one-carbon metabolism and leads to increased homocysteine, and an aberrant methyl pool, which may in turn affect LINE-1 methylation.
LINE-1 is a non-terminal repeated sequence, with independent transposition functions. The transposition activity of LINE-1 is controlled by epigenetic regulation, particularly DNA methylation [Iskow et al., 2010]. Some studies about tumors have indicated that LINE-1 hypomethylation leads to increased LINE-1 retrotransposition, thereby generating abnormal inserts [Cruickshanks and Tufarelli, 2009; Iskow et al., 2010] that can act as mutagens [Belancio et al., 2008; Goodier and Kazazian, 2008], affect gene expression [Perepelitsa-Belancio and Deininger, 2003; Han et al., 2004], and be required to silence chromosome X by Xist [Chow et al., 2010]. Therefore, transposition activity changes can affect the structure and function of the genome [Belancio et al., 2008]. In this study, we focused on the LINE-1 5′UTR to conduct our methylation analysis. It is known that the 5′UTR has an internal RNA polymerase II promoter that directs transcription from the 5′ end of the element [Swergold, 1990], and it also contains cis-acting binding sites for multiple transcription factors [Minakami et al., 1992; Tchenio et al., 2000; Yang et al., 2003; Kuwabara et al., 2009]. Some studies show that hypomethylation of the LINE-1 5′UTR also occurs in malignant cells and cancerous tissues, and is correlated with an increase in LINE-1 mRNA and/or ORF1p expression [Alves et al., 1996; Goodier and Kazazian, 2008]. These observations suggest that aberrant LINE-1 methylation in mouse ESCs may lead to novel transposition activity, thus affecting early embryonic development.
Folate is regarded as essential for DNA methylation, and also has important roles in de novo purine and pyrimidine biosynthesis [Kim et al., 2009]. Therefore, in this study, we simultaneously evaluated mESC morphology, proliferation and apoptosis in folate-deficient conditions, no change was found in cell morphology among groups and no significant differences were built in proliferation and apoptosis (Fig. 6). The reason may be due to the trace folate of FBS to meet the needs of the cell growth, while the changes of intracellular methylation level may be more sensitive to folate deficiency. Therefore, in the short term, we found only methylation changes, without affecting cell proliferation. Interestingly, we found that SSEA-1 expression gradually reduced in FN, FD, and FF from day 9, as indicated by flow cytometry and immunofluorescent staining. SSEA-1 is a specific marker of mouse ESCs [Muramatsu and Muramatsu, 2004]. Studies indicate that SSEA-1 is involved in cell–cell interactions [Kudo et al., 1998] and participates in cell adhesion and signal transduction during early embryonic development [Staudacher et al., 1999; Becker and Lowe, 2003]. Novel SSEA-1 expression in our study indicates that the features of the mouse ESCs may have changed presumably by affecting cell adhesion or signal transduction, but the specific effect of SSEA-1 on mouse ESCs is still unclear and needs further study.
ESCs have the potency to develop into any type of cell. Therefore, our study focused on whether folate deficiency had an effect on cell differentiation. Recent studies have established the fundament roles of several transcription factors, namely Oct4, Sox2, and Nanog, in the self-renewal and pluripotency of mouse ESCs. These factors maintain their own and each other's transcription and activation by combinatorial interactions with genes responsible for the ESC phenotype, while repressing genes required for differentiation [Niwa et al., 2000; Chambers et al., 2003; Chambers and Smith, 2004; Masui et al., 2007]. In the study, we assayed the expression of Oct4, Sox2, and Nanog by real-time PCR, and found no significant expression differences of these genes in all groups. However, there was an increase in FD compared with that in FN, and a sharp increase in FF at day 18. Previous studies have found that folate deficiency can cause reduced genome methylation accompanied by particular gene methylation changes that affect their expression [Jjingo et al., 2012]. Thus, we speculated that novel expression of Oct4, Sox2, and Nanog may be increased by different methylation levels in folate-deficient condition. In future study, we will extend the duration of treatments, and induce ESC differentiation to the embryoid bodies (EB) stage to conduct a folate treatment study combined with LINE-1 function experiments to investigate the effects on cell function.
In summary, our study found that low folate culture causes intracellular folate insufficiency and homocysteine accumulation in mouse ESCs, and leads to hypomethylation of LINE-1, indicating that one-carbon metabolism disorder in ESCs directly leads to reduced LINE-1 methylation levels. In addition, SSEA-1 of mouse ESCs expression is decreased in folate-deficient condition, suggesting that folate deficiency may affect the characteristics of mouse ESCs, which requires further study. This study provides preliminary evidence of folate deficiency affecting early embryonic development.




