Abstract
No ideal serum biomarker currently exists for the early diagnosis of colorectal cancer (CRC). Magnetic bead-based fractionation coupled with MALDI-TOF MS was used to screen serum samples from CRC patients, healthy controls, and other cancer patients. A diagnostic model with five proteomic features (m/z 1778.97, 1866.16, 1934.65, 2022.46, and 4588.53) was generated using Fisher algorithm with best performance. The Fisher-based model could discriminate CRC patients from the controls with 100% (46/46) sensitivity and 100% (35/35) specificity in the training set, 95.6% (43/45) sensitivity and 83.3% (35/42) specificity in the test set. We further validated the model with 94.4% (254/269) sensitivity and 75.5% (83/110) specificity in the external independent group. In other cancers group, the Fisher-based model classified 25 of 46 samples (54.3%) as positive and the other 21 as negative. With FT-ICR-MS, the proteomic features of m/z 1778.97, 1866.16, 1934.65, and 2022.46, of which intensities decreased significantly in CRC, were identified as fragments of complement C3f. Therefore, the Fisher-based model containing five proteomic features was able to effectively differentiate CRC patients from healthy controls and other cancers with a high sensitivity and specificity, and may be CRC-specific. Serum complement C3f, which was significantly decreased in CRC group, may be relevant to the incidence of CRC. J. Cell. Biochem. 114: 448–455, 2013. © 2012 Wiley Periodicals, Inc.
Abbreviations:
CRC, colorectal cancer; ROC, receiver operating characteristic; CEA, carcinoembryonic antigen.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, and it is the second leading cause of cancer deaths in the United States [Jemal et al., 2010, 2011]. Both the incidence and mortality of CRC have declined over the last two decades, which has been attributed to the early detection and treatment of adenomas and CRC by the American Cancer Society [Jemal et al., 2010]. Colonoscopies, which are currently the gold standard for the early detection of CRC, are invasive and uncomfortable, and they require professionally trained staff [Rozen, 2004]. Although serum carcinoembryonic antigen (CEA) has been widely used for CRC, its lack of specificity and sensitivity preclude the use of CEA for the early detection of CRC [Duffy et al., 2007]. Therefore, the discovery of more accurate and reliable biomarkers for the early diagnosis of CRC remains an urgent need.
Advances in clinical proteomics, which uses mass spectrometry (MS)-based protein profiles of easily accessible body fluids to distinguish different patients, may offer a solution to this problem [Wulfkuhle et al., 2003]. Though so far a number of CRC-associated tissue proteins have been discovered in multiple studies, with the greater majority being 2D gel-based discoveries coupled to MS/MS, only a limited number of them have been validated in serum for non-invasive testing for CRC [Jimenez et al., 2010]. In particular, the use of magnetic-bead fractionation-based analyses, which have been applied to the early detection of oral cancer and head and neck cancer, appears to have significant potential for the discovery of biomarkers [Cheng et al., 2005; Freed et al., 2008].
In the present study, we present the results of a study that used magnetic bead-based fractionation coupled to matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) for the analysis of serum from CRC patients, healthy controls, and other cancer patients. The identified peptide markers described here could aid in the early diagnosis of CRC.
METHODS
Study Overview
The flowchart of the study is shown in Figure 1. The comparison among CRC patients and healthy controls indicated that the intensities of 25 proteomic features were statistically different (P < 0.001 and average mass area > 25.0). Then individual comparisons of different stage CRC group and healthy controls showed that there were six common proteomic features. Further we built diagnosis models between CRC and healthy controls using the above five overlapped proteomic features with different algorithms. Finally, the Fisher-based class prediction was chosen to carry out with best performance. And differential proteomic features between CRC and healthy controls were identified using Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS). This trial was registered on ClinicalTrial.gov (NCT 01604798).
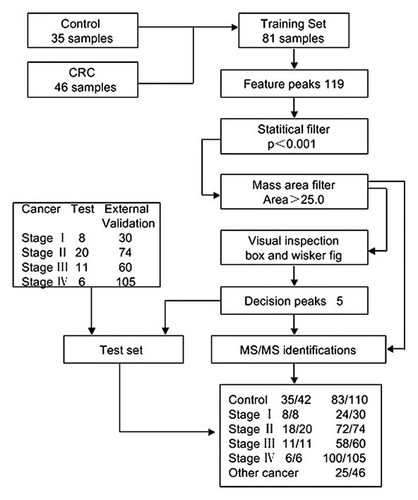
Flow chart of the study The diagram shows the approach used for development and validation of the model with five proteomic features.
Serum Samples Collection
All of the cancer serum samples were obtained from patients with histologically confirmed cancer or leukemia in Zhongshan Hospital, Fudan University, China. The healthy control samples were collected from healthy volunteers. All of the patients and volunteers provided written informed consent. This study was approved by the Institutional Review Board of Medical College, Fudan University.
All fasting blood samples were prepared without anticoagulant and left to clot at room temperature for 1.5 h. The serum was then isolated by centrifugation at 3,000g for 10 min at room temperature and stored at −80°C. All samples were subjected to one freeze–thaw cycle.
Peptide Extraction and MALDI-TOF MS Analysis
Prior to MS analysis, the serum samples were fractionated using immobilized metal affinity chromatography-copper (IMAC-Cu) magnetic beads from the National Center of Biomedical Analysis (NCBA) according to the manufacturer's recommendations [Wang et al., 2007]. Briefly, 5 µl of bead suspension and 20 µl of binding solution were mixed with 5 µl of serum before incubating for 10 min. During all subsequent washing steps, a magnetic separator was utilized to keep all of the beads together with the bound protein fraction in one location within the tube. To remove the unbound proteins, the beads were washed three times with 100 µl of wash solution. The bound peptides were then eluted using 20 µl of elution solution. Finally, 1 µl of the protein solution was mixed with 1 µl of α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution and was spotted onto a 600-µm spot of an MTP384 target plate (Bruker Daltonics, Germany). Air-dried targets were analyzed within 2 h using an UltraFlex III MALDI-TOF MS (Bruker Daltonics). Instrument calibration parameters were determined using standard peptide and protein mixtures. All measurements were performed in a blind manner.
Data Analysis
Mass spectra were subjected to curve smoothing, baseline subtraction and peak labeling using FlexAnalysis 3.0 software (Bruker Daltonics), where all quality peaks (signal-to-noise ratio > 5) with m/z values between 800 and 10,000 Da were compiled and labeled. Peaks from different spectra were aligned with the criteria that m/z values from two spectra within 0.1% were considered to represent the same peptide. Next, BioExplorer™ (Bioyong Tech, Beijing, China) was used to compile the peaks across the spectra, apply different algorithms to generate models for class prediction and then validate the sensitivity and specificity with two sets of independent serum samples. SPSS version 16.0 (SPSS, Inc., Chicago, IL) was used to construct Box-and-whiskers plots.
Peptide Sequencing
The sequences of differential peptides between CRC and healthy controls were identified using an on-line nanoLC-MS/MS system, which was Agilent 1100 series HPLC system (Agilent) coupled to a Apex-Qe FT-ICR-MS (Bruker Daltonics). The extracted peptides by magnetic beads were desalted and sequenced by MS/MS mode. The selected ion was fragmented by collision-induced dissociation. The MS/MS data were processed and submitted to the search engine Mascot (www.matrixscience.com). Peptide mass tolerance was set at 10 ppm, fragment ion mass tolerance was set at 0.01 Da, and the mass type of parent peptide and peptide fragment were set at monoisotopic.
RESULTS
In the present study, a total of 46 serum samples from the CRC patients and 35 from healthy controls were assigned to the training set, and 45 from CRC and 42 from controls were used as the test set. Then we further chose 269 from CRC and 110 from controls as external validation set. In addition, we validated the model with 46 from other solid cancer and leukemia group to evaluate whether the model was CRC-specific. The other cancers group includes 10 patients with hepatocellular cancer, 8 with gastric cancer, 10 with lung cancer, 8 with breast cancer, and 10 with leukemia (including 4 acute lymphocytic leukemia, 2 acute myeloid leukemia-M2, and 4 acute promyelocytic leukemia according to French-American-British classification criteria). The clinical characteristics of cancer patients and healthy controls are shown in Table I.
| Training set | Test set | External validation set | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC (n = 46) | Control (n = 35) | CRC (n = 45) | Control (n = 42) | CRC (n = 269) | Control (n = 110) | Hepatocellular cancer (n = 10) | Gastric cancer (n = 8) | Lung cancer (n = 10) | Breast cancer (n = 8) | Leukemia (n = 10) | |
| Gender (M:F) | 27:19 | 19:16 | 25:20 | 24:18 | 145:124 | 64:46 | 6:4 | 3:5 | 6:4 | 0:8 | 5:5 |
| Age (year), median (range) | 62.0 (32–80) | 59.0 (41–79) | 60.0 (37–74) | 61.0 (35–76) | 58.0 (25–79) | 51.0 (24–78) | 53.0 (40–70) | 48.0 (41–75) | 46.5 (36–65) | 40.0 (28–60) | 35.0 (20–50) |
| Stage I | 4 | 8 | 30 | 2 | 0 | 1 | 0 | ALL: 4 | |||
| Stage II | 16 | 20 | 74 | 5 | 4 | 6 | 5 | AML-M2: 2 | |||
| Stage III | 13 | 11 | 60 | 3 | 4 | 3 | 2 | APL: 4 | |||
| Stage IV | 13 | 6 | 105 | 0 | 0 | 0 | 1 | ||||
- CRC, colorectal cancer; M, male; F: female; CEA, carcinoembryonic antigen; ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; APL, acute promyelocytic leukemia.
To evaluate the reproducibility of the assay, we used one pooled serum sample from 8 CRC patients to analyze 6 within-run assays and 6 between-run assays. The mean coefficient of variation (CV) of the within-run assays was 16.1% (8.1–27.8%), and the mean CV of the between-run assays was 18.9% (4.8–27.0%; Table II).
| m/z | Within-run assays | Between-run assays | ||
|---|---|---|---|---|
| MRI (%) | CV (%) | MRI (%) | CV (%) | |
| 1945.58 | 11.0 | 12.7 | 17.2 | 21.6 |
| 2990.99 | 11.5 | 14.6 | 17.0 | 27.0 |
| 3315.53 | 16.0 | 10.5 | 18.4 | 26.5 |
| 4153.83 | 26.8 | 27.8 | 22.9 | 12.9 |
| 6436.94 | 26.7 | 8.1 | 27.0 | 4.8 |
| 8929.61 | 10.6 | 22.9 | 8.4 | 20.7 |
- MRI, mean relative intensity; CV, coefficient of variation.
MALDI-TOF MS analysis on the fractionated serum samples resolved a total of 119 peaks that ranged from 800 to 10,000 Da, of which 38 were with P value < 0.001 (t-test) and 25 with average mass area > 25.0 (Fig. 2 and Table III). And individual comparisons of different stage CRC group and controls indicated that there were six common proteomic features (m/z 1778.72, 1865.90, 1934.79, 2022.15, 4587.64, and 9380.49) with significant difference between CRC patients and controls. Then five overlapped proteomic features (m/z 1778.97, 1866.16, 1934.65, 2022.46, and 4588.53) were selected (Table IV). All of five selected proteomic features were of lower intensity in CRC compared with controls (Fig. 3A). And then, the five proteomic features were further taken for unsupervised hierachical clustering analysis and also showed significant difference between CRC patients and controls (Fig. 3B).
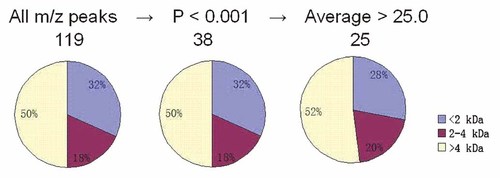
Proteomic feature selection of serum peptide profiling data The peak list was subjected to a Mann–Whitney's U-test for CRC versus controls. Only peaks with adjusted P-values of <0.001 were passed through a second filter (average peak area > 25.0).
| m/z | MRI (SD) in CRC group | MRI (SD) in control group | P-value | Peptide sequence | Peptide name |
|---|---|---|---|---|---|
| 1480.53 | 38.73 (47.6) | 11.18 (2.01) | 9.51E−04 | R.SGPFGQIFRPDNF.V | Tubulin beta-2C chain |
| 1778.84 | 13.67 (7.54) | 42.19 (16.84) | <1.00E−06 | S.SKITHRIHWESASLL.R | Complement C3f |
| 1866.03 | 20.08 (17.1) | 153.18 (78.3) | <1.00E−06 | R.SSKITHRIHWESASLL.R | Complement C3f |
| 1934.93 | 8.67 (2.63) | 25.05 (13.67) | <1.00E−06 | S.SKITHRIHWESASLLR.S | Complement C3f |
| 2022.27 | 31.68 (16.26) | 144.39 (104.06) | 2.38E−06 | R.SSKITHRIHWESASLLR.S | Complement C3f |
| 2553.92 | 33.29 (20.26) | 21.23 (4.8) | 9.49E−04 | K.SSSYSKQFTSSTSYNRGDSTFES.K | FGA isoform 1 of fibrinogen alpha chain precursor |
| 3216.48 | 49.79 (20.26) | 71.24 (16.78) | 5.54E−06 | R.HGFESGDFVSFSEVQGMVELNGNQPMEIK.V | Ubiquitin-like modifier activating enzyme 1 |
| 3315.74 | 105.73 (40.63) | 178.35 (25.23) | <1.00E−06 | R.FLGDRDFNQFSSGEKNIFLASFVHEYSR.R | Alpha-fetoprotein precursor |
| 3883.77 | 26.68 (10.07) | 34.32 (5.73) | 2.21E−04 | R.SARLNSQRLVFNRPFLMFIVDNNILFLGKVNRP.-M.SIPPEVKFNKPFVFLMIE | Plasma serine protease inhibitor precursor |
| 4136.23 | 45.59 (19.79) | 31.55 (10.09) | 3.47E−04 | QNTKSPLFMGKVVNPTQK - R.TIHLTMPQLVLQGSYD | PRO2275 |
| 4269.67 | 30.64 (13.2) | 19.73 (6.22) | 3.58E−05 | QDLLAQAELPAILHTELNLQK.L L.SALVETRTIVRFNRPFL | Angiotensinogen precursor |
| 4627.64 | 66.05 (27.62) | 85.35 (17.91) | 9.49E−04 | MIIVPTDTQNIFFMSKVTNPKQA | Alpha-1-antichymotrypsin precursor |
- CRC, colorectal cancer; MRI, mean relative intensity; SD, standard deviation.
| m/z | Median mass area | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Control group | Stage I CRC group | Stage II CRC group | Stage III CRC group | Stage IV CRC group | Stage I CRC vs. control | Stage II CRC vs. control | Stage III CRC vs. control | Stage IV CRC vs. control | |
| 1778.97 | 44.86 | 12.27 | 16.76 | 15.69 | 13.09 | 1.71E−02 | <1.00E−06 | <1.00E−06 | <1.00E−06 |
| 1866.16 | 134.83 | 12.96 | 19.01 | 16.29 | 16.22 | 5.16E−03 | <1.00E−06 | <1.00E−06 | <1.00E−06 |
| 1934.65 | 26.00 | 8.53 | 10.20 | 9.14 | 9.49 | 1.88E−06 | 1.46E−06 | <1.00E−06 | 1.39E−06 |
| 2022.46 | 119.48 | 21.53 | 32.04 | 32.39 | 27.47 | 1.88E−06 | 5.65E−06 | 5.01E−06 | 2.28E−06 |
| 4588.53 | 41.63 | 12.31 | 17.59 | 14.61 | 20.56 | 7.39E−03 | 1.27E−05 | <1.00E−06 | <1.00E−06 |
- CRC, colorectal cancer; m/z, mass-to-charge ratio.
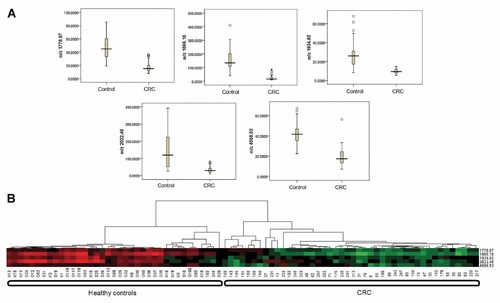
A,B: Distribution of proteomic features in CRC group and controls (A) Intensity distribution of five selected peaks between the controls and CRC patients are shown in a box-and-whisker diagram. The line in the box marks the median, the central rectangle spans the first quartile to the third quartile, and the whiskers above and below the box show the locations of the maxima and minima. The open dots indicate extreme outliers. B: Clustering analysis of five selected peaks in their distribution among CRC and control samples. The intensities arrangement of the five peaks in 81 samples in binary format was by unsupervised, average-linkage hierarchical clustering using standard correlation as a distance metrics between CRC group and controls. Columns represent samples, rows are m/z peaks as indicated by the average molecular weight. The heat map scale of normalized ion intensities is from −1 (green) to +1 (red) with the midpoint at 0 (black).
Therefore, the models with the five peptide peaks were generated using different algorithms. Then the performances of the models were detected with the training and test sets. Of the four models, the Fisher-based model showed best performance, which could discriminate CRC patients from healthy controls with 100% sensitivity and 100% specificity in the training set, and 95.6% sensitivity and 83.3% specificity in the test set (Tables V and VI). Additionally, with the ROC curves, the Fisher-based model had a higher AUC (1.000; 95% CI, 1.000–1.000) than CEA alone (0.721; 95% CI, 0.611–0.820), which exhibited a higher classification performance in the training set (Fig. 4).
| Algorithm | Training set | Test set | ||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| SVM | 100% (46/46) | 100% (35/35) | 84.4% (38/45) | 78.6% (33/42) |
| KNN | 100% (46/46) | 100% (35/35) | 86.7% (39/45) | 78.6% (33/42) |
| Fisher | 100% (46/46) | 100% (35/35) | 95.6% (43/45) | 83.3% (35/42) |
| RBF | 100% (46/46) | 100% (35/35) | 88.9% (40/45) | 76.2% (32/42) |
- SVM, support vector machine; KNN, k-nearest neighbor, RBF, radial basis function neural network.
| Training set | Test set | External validation set | |
|---|---|---|---|
| Sensitivity | 100% (46/46) | 95.6% (43/45) | 94.4% (254/269) |
| Specificity | 100% (35/35) | 83.3% (35/42) | 75.5% (83/110) |
| Positive predictive value | 100% (46/46) | 86% (43/50) | 90.4% (254/281) |
| Negative predictive value | 100% (35/35) | 94.6% (35/37) | 84.7% (83/98) |
| Accuracy | 100% (81/81) | 89.7% (78/87) | 88.9% (337/379) |
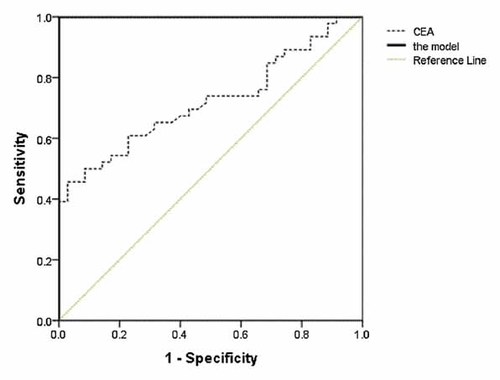
ROC curves of the model and CEA in the training set performance of the model (solid line) and CEA (dotted line) were shown in ROC space. Gray line indicates the reference line.
To evaluate the robustness of the Fisher-based model, we further tested the peptide signatures on an external independent set of 269 serum samples from CRC patients and 110 from healthy controls. None of the samples had been previously included in the former analysis, which therefore allowed for the estimation of true diagnosis accuracy. Finally, we obtained 94.4% (254/269) sensitivity, 75.5% (83/110) specificity, and 88.9% (337/379) accuracy, respectively (Table VI). In addition, the Fisher-based model classified 25 of 46 samples (54.3%) as cancer and the remaining 21 (45.7%) as controls in other cancers group.
With LC-MS/MS detection, 12 of 25 differential peptides between CRC and controls were identified successfully (Table III). After database searching, the peptide of m/z 1778.97, 1866.16, 1934.65, and 2022.46 were all identified as fragments of complement C3f.
DISCUSSION
We directly profiled protein and peptide patterns from magnetic bead-fractionated serum samples using MALDI-TOF MS, and determined several markers that differentiated CRC patients from healthy controls with a high sensitivity and specificity. In addition, the model only correctly classified almost half of samples in other cancers group. The intensities of the proteomic feature m/z 1778.97, 1866.16, 1934.65, and 2022.46, which were identified as fragments of complement C3f, were decreased in the serum samples from the CRC patients compared with healthy controls.
CRC is one of the leading causes of cancer-related death worldwide, and early diagnosis of CRC allows for more effective treatments that could improve the long-term survival. No current methods (e.g., colonoscopies or serum CEA test) have been established as well-accepted tools for the early diagnosis of CRC because of low-adherence rates, high costs or low sensitivity [Duffy, 2001]. Thus, improved and innovative methods with high performance for early CRC detection are urgently needed.
To address the above limitations, we aimed to discover accurate and reliable serum biomarkers that distinguish CRC from controls. Due to the heterogeneous character of CRC, a single biomarker is not likely to provide sufficient diagnostic power. Instead, a panel of multimarker assays should be developed to reach diagnostic accuracy. Advances in clinical MS-based proteomics that focus on modifications of the proteome in the presence of disease offer the potential to discover the much-needed biomarkers [Hanash et al., 2011]. Serum proteomics focuses on low-molecular-weight peptides that are believed to be tumor expressed and host response proteins, and it reflects the biological states of altered cells as tissue leakage proteins [Anderson and Anderson, 2002; Rosenblatt et al., 2004; de Noo et al., 2006].
In the past decade, surface-enhanced laser desorption/ionization-time-of-flight mass spectrometry (SELDI-TOF MS), MALDI-TOF MS, LC-MS, and other quantification methods have been used for the expression analysis of low-molecular-weight serum proteins [Jimenez et al., 2010]. Habermann et al. [2006] screened sera from 58 CRC patients and 32 healthy controls for potential differences using SELDI-TOF MS, and identified that the most prominent m/z values revealed a member of the complement system, the stable form of C3a anaphylatoxin, which was then validated in independent sample sets with a sensitivity of 96.8% and a specificity of 96.2% using a specific enzyme-linked immunosorbent assay. Several studies regarding the clinical applicability of SELDI-TOF have also established promising prospective models and identified certain proteins (e.g., APOC1, C3a, and HNP1) as biomarkers for the early diagnosis of CRC [Gemoll et al., 2010]. However, there are some inherent drawbacks with the SELDI-TOF technology, such as the inability for direct identification, poor resolution and mass accuracy and low reproducibility [Wang et al., 2009]. Therefore, magnetic bead affinity purification for serum protein fractionation, followed by MALDI-TOF MS with its high accuracy and reproducibility, has been used to identify certain proteins associated with gastric cancer and bladder cancer [Ebert et al., 2006; Schwamborn et al., 2009]. In the present study, we built the Fisher-based model for early CRC detection with a high-classifying performance using this technology, and then observed that decreased levels of complement C3f were associated with the incidence of CRC, but not with other solid cancers or hematological malignancies.
In serum proteomics studies, biological variations, pre-analytical variations, and analytical reproducibility are all possible confounding factors [Albrethsen, 2007]. Therefore, to improve analytical performance, we chose healthy controls of equivalent age and gender distributions; standardized the sample collection, storage and fractionation protocols; applied quality control samples and optimized the parameters of the MALDI-TOF MS instrument. The CV values of the within-run and between-run assays in our study were acceptable, which confirmed the usefulness of our Fisher-based model.
Our Fisher-based model with five peptide peaks was sufficient to correctly classify 100% of CRC patients and 100% of healthy controls in the training set, 95.6% of CRC patients and 83.3% of healthy controls in the test set, and 94.4% of CRC patients and 75.5% of healthy controls in the external validation set. In addition, the Fisher-based model only classified almost half of samples from other cancers group as cancer. Therefore, the Fisher-based model had a much higher performance than CEA, which is widely used clinical marker, and was more suitable for the diagnosis of CRC than other cancers. Further characterization of differential proteomic features may provide direct insights into cancer pathogenesis, which could further enable us to develop immunoassay measurements of these potential markers. Of note four of five peptides from the Fisher-based model (m/z 1778.97, 1866.16, 1934.65, and 2022.46) were identified as fragments of complement C3f, which significantly decreased in CRC group.
The changes of complement C3f have been reported to be associated with various cancers, including hepatocellular cancer, nasopharyngeal cancer, and adult T-cell leukemia, which indicates that it serves important function but may decrease the applicability as a specific marker [Chang et al., 2006; Ishida et al., 2008; An et al., 2010]. However, the spectrum of specific fragments of the proteins may be cancer type-specific [Villanueva et al., 2006]. And our results also indicated that the Fisher-based model was more suitable for the diagnosis of CRC than other cancers.
The complement system is a major mediator of immune system against tumors, and complement C3 plays a central role in the activation of complement system through all three pathways (classical, alternative, and lectin) [Sahu and Lambris, 2001]. Ajona et al. [2004] demonstrated that most non-small cell lung cancer cell lines highly expressed factor H, an inhibitor of complement activation, and decreased the susceptibility of these cells to complement-mediated cytotoxicity. In the present study, fragments of complement C3f were significantly decreased in CRC group. Complement C3f is a byproduct of C3b, the activated forms of C3, after it has been cleaved to iC3b. We thought that there was immune escape of cancer, and decrease of complement C3f may be attributed to activity change of the enzyme responsible for cleavage of C3b (e.g., factor I, cellular membrane type-1 matrix metalloproteinase) [Rozanov et al., 2004; Okroj et al., 2008]. There was another possibility that exoprotease activities superimposed on the ex vivo coagulation and complement-degradation pathways contribute to generation of not only cancer-specific but also cancer type-specific serum peptides [Villanueva et al., 2006, 2008; Huijbers et al., 2010]. In addition, some studies indicate that the cause of decrease of complement C3f is the pathogens of infections in cancer patients, and the complement system could be activated by the pathogens to defend the body itself against infection and simultaneously release anaphylatoxins to activate inflammatory cells [Liang et al., 2010]. The mechanism that how the enzymes or the complement and other systems contribute to the observed differences in serum peptide patterns remains little understood and requires further study.
In the present study, we only used IMAC-Cu magnetic beads to fractionate the serum proteins and peptides. Other magnetic bead-based affinity surfaces (e.g., weak cationic exchange and hydrophobic) could also be used to produce discriminatory protein peaks that could be combined with our Fisher-based model.
CONCLUSION
We used IMAC-Cu magnetic bead fractionation to purify serum proteins prior to MALDI-TOF MS analysis, and developed the Fisher-based model of five proteomic features with a high sensitivity and specificity for the early diagnosis of CRC, which may be CRC-specific. Serum complement C3f, levels were significantly decreased in CRC group, may be relevant to the incidence of CRC.
Acknowledgements
This work was supported by Key Projects of the Clinical Disciplines, which is administered by the Ministry of Health (Projects from 2010–2012), the National Natural Science Foundation (30973416, 81101566), and the Talent Fund of Shanghai Municipal Health Bureau (XYQ2011017, XBR2011031).




