Glutamate-ammonia ligase and reduction of G0 population in PANC-1 cells
Abstract
In our previous study, we screened and isolated genes that were up-regulated after partial pancreatectomy using transcriptomic analysis and glutamate-ammonia ligase (GLUL) was selected for further study based on its effect on differentiation and proliferation. In the immunohistochemical analysis, GLUL was highly up-regulated in the acinar cells and the ductal cells in the pancreas damaged through partial pancreatectomy. Overexpression of GLUL enhanced the proliferation of PANC-1 cells and INS-1 cells. GLUL overexpression shifted the major population of PANC-1 cells from the G0/G1 phase to G2/M phase. In the double thymidine blocking analysis, similar cycle duration was observed between mock cells and GLUL-overexpressing cells while GLUL-overexpressing cells were partially resistant to thymidine blocking. In the FACS analysis of cells stained with Pyronin Y and Hoechst 33342, GLUL-overexpressing cells showed lower population of cells in the G0-quiescent phase than mock cells (5–12%). In addition, GLUL-overexpressing cells had high activation levels of AKT, ERK1/2, JNK, PCNA, c-FOS, and P70S6K in PANC-1 cells. Taken together, these results suggest that GLUL contributes to pancreatic regeneration. J. Cell. Biochem. 114: 303–313, 2013. © 2012 Wiley Periodicals, Inc.
Injury produces a functional or structural loss of tissue and stimulates inflammation. As a protective response, inflammation destroys parts of the damaged region and initiates the healing process. These healing processes are mediated by the secretion of proteins from local tissues and immune cells. Some pro-inflammatory cytokines such as IL-1β, IFN-γ, and TNF-α inhibit proliferative pathways [Blandino-Rosano et al., 2008] and other factors induced by inflammation protect the tissue from the damaging signals [Savković et al., 2007]. The liver, skin, and kidney are good examples of tissues that have developed processes to control the balance between inflammation and healing processes [Iredale, 2007; Goligorsky, 2008; Eming et al., 2009].
The same phenomenon can be found in the pancreas. Acute or chronic inflammation induced cell proliferation in the pancreas [Strobel et al., 2007] and immunosuppressants and inhibit pancreatic regeneration [Nir et al., 2007]. Pancreatitis has been shown to activate hedgehog components which supplement tissue regeneration [Fendrich et al., 2008], and transforming growth factor β which regulates islet plasticity in the pancreas [Hanley and Rosenberg, 2007]. Partial pancreatectomy, the dissection of the pancreas, is an injury that induces inflammation and regeneration. Regeneration studies following partial pancreatectomy have been reported in animals and humans [Hayashi et al., 2003; Berrocal et al., 2005; Peshavaria et al., 2006]. It has been reported that partial pancreatectomy can upregulate well-known transcription factors of insulin [Liu et al., 2007] or putative regeneration factors. For example, clusterin (CLU) was shown to have regenerating function in a partial pancreatectomy model [Min et al., 2003; Kim et al., 2006].
Glutamate-ammonia ligase (GLUL) is the enzyme that combines glutamate and ammonium ion into glutamine [Eisenberg et al., 2000] and its activity has been shown to be high in the liver, brain, kidney, spleen, and testis [Lund, 1970]. The expression of GLUL was related to cell proliferation in the liver [Wu, 1984], intestinal epithelium [DeMarco et al., 1999], retina [Kase et al., 2006], and skin [Vermeulen et al., 2008]. In the annelid, the regeneration of the whole body seems to be regulated by GLUL [Myohara et al., 2006; Niva et al., 2008]. The expression of GLUL was regulated by extracellular hormones [Blutstein et al., 2006; Wang and Watford, 2007] and intracellular glutamine [Stanulović et al., 2006; Huang et al., 2007]. In addition, the activity of GLUL has been shown to be restricted by a glutamine analogue [Lejczak et al., 1981]. The transcription efficiency of Glul was high in the pancreas; but, its protein expression was not detected in the pancreas [Straaten et al., 2006]. WNT signal seems to regulate the expression of GLUL in the pancreatic neoplasm [Audard et al., 2008]. Glutamine is the major carrier of nitrogen, which is a biosynthetic resource of nucleotide and hexoamine, an alternative supplier of the TCA cycle, and a triggering factor of ERK and PI3K signals [DeBerardinis and Cheng, 2010].
Recently, we reported that our subtractive hybridization screening in sham-operated pancreas tissue showed that Glul was up-regulated in the pancreas by partial pancreatectomy and regulated several transcription factors involved in pancreatic regeneration [Choi et al., 2010]. In addition, GLUL gene expression was increased during pheochromocytoma cell growth and proliferation [Choi et al., 2010]. However, specific physiological functions of GLUL in partial pancreatectomy remain unclear. Based on our study, we analyzed the localization of GLUL in the pancreas tissue after partial pancreatectomy, compared the effects of GLUL in two pancreatic cell lines, and analyzed the effect of GLUL on the cell cycle in this study.
MATERIALS AND METHODS
Cell Culture, DNA Constructs, and Antibodies
The 293T and PANC-1 (ATCC CRL-1469) cells were cultured in DMEM medium (Gibco-BRL, Grand Island, NY) with 10% FBS (Gibco-BRL) and penicillin/streptomycin (Gibco-BRL). INS-1 cells were cultured in RPMI 1640 medium (Gibco-BRL) with 10% FBS, 11.1 mM glucose and penicillin/streptomycin. Media were changed every 2 days. As an inhibitor of GLUL, L-methionine-D,L-sulphoximine (MSO; Sigma, St. Louis, MO) was used at the concentration of 5 or 15 mM.
Complete coding sequences of Glul (NM_017073) from rat pancreas cDNA was cloned into pGEM-T Easy vector (Promega, Madison, WI) and moved into modified pMSCVhyg (antigenic-myc sequence is inserted at multi-cloning site; Clonetech, Mountain View, CA) expression vector. shRNA of Glul was designed with Insert Design Tool for the pSilencer Vectors (Applied Biosystems, Austin, TX). The top (5′-GAT CCA CCT CCA ACA TCA ACG ACT TTC AAG AGA AGT CGT TGA TGT TGG AGG TTT TTT TGG AAA-3′) and bottom (5′-AGC TTT TCC AAA AAA ACC TCC AAC ATC AAC GAC TTC TCT TGA AAG TCG TTG ATG TTG GAG GTG-3′) strands of insert were heated to 90°C for 3 min and annealed in 37°C for 1 h. The product was integrated to modified pMSCVhyg (H1 promoter was inserted to multi-cloning site) with BamHI and HindIII (New England BioLabs, Beverly, MA). These target sequences are homologous between human and rat.
Antibodies were used as follows: anti-GLUL (BD Bioscience, Franklin Lakes, NJ), anti-c-MYC (9E10), anti-Ki-67 (Chemicon, Temecula, CA), biotinylated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and horseradish peroxidase (HRP)-conjugated secondary antibody (KPL, Gaithersburg, MD).
Partial Pancreatectomy
Eight-week-old male Sprague-Dawley rats were anesthetized by i.m. injection of 40 mg/kg of ketamine hydrochloride (Huons, Seoul, Korea) and 5 mg/kg xylazine hydrochloride (Rompun; Bayer Korea, Seoul, Korea). Their abdomens were opened through linea alba. For the 60% partial pancreatectomy, the splenic portion was removed from the pancreas. For the sham operation, the abdomen was opened and the pancreas was left intact. The incision was closed using 4–0 silk thread. After 2 days, the remaining pancreas from two groups was homogenized in liquid N2. A part of the sample was treated with TRIzol™ (Invitrogen, Carlsbad, CA) to synthesize cDNA. Another part was treated with chilled NP-40 lysis buffer containing a protease inhibitor cocktail (Sigma) and centrifuged to obtain a clear protein soup. The other part was fixed with paraformaldehyde solution for the tissue staining. Samples were stored in −70°C. All animal procedures were approved by Institutional Animal Care and Use Committee at CHA University as required (Project Number: IACUC090016).
Immunohistochemistry
The mouse pancreas was embedded in paraffin, sectioned at a thickness 5 µm, and fixed in 4% formaldehyde overnight at 4°C. For antibody staining, the sections were deparaffinized and placed into a 3% solution of hydrogen peroxide for 5 min. And these were washed twice with phosphate-buffered saline with Tween 20 (PBST; 133 mM NaCl, 10 mM sodium potassium phosphate, pH 7.4 containing 0.1% Tween 20 [Bio-Rad, Hercules, CA]), placed in a blocking solution with 0.5% bovine serum albumin/PBST for 30 min. After incubation with the primary antibody for 1 h followed by washing, biotinylated secondary antibody was applied for 30 min and streptavidin-horseradish peroxidase substrate, and counter staining was performed with hematoxylin.
Immunocytochemistry
Cells were fixed with 70% ethanol in PBS for overnight at 4°C, permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 10 min, and stained with the primary antibody. Alexa Fluor-conjugated IgG (Invitrogen) was used for the secondary antibody. Nuclei were stained with Hoechst 33342 (Sigma).
Viral Production and Transduction
Three micrograms of pMSCVhyg vector, 3 µg of pVPack-VSV-G (Stratagene, La Jolla, CA), and 3 µg of pVPack-GP (Stratagene) were mixed with 600 µl of 150 mM NaCl and 150 µl of 10 mM PEI (Polyscience, Warrington, PA). The transfection mixture was incubated for 15 min at room temperature and added to 70% confluent 293T cells in a 100Φ dish containing 5.4 ml of normal media. Media were changed after 5 h of PEI treatment. After 2 days, soup from the packaging cell were collected and filtrated. Viral particles were concentrated at 50,000 g for 2 h. Pellet was suspended in culture media with 8 µg/ml polybrene (Invitrogen). Viral media were treated to target cells for 1 day.
Reverse Transcriptase-Polymerase Chain Reaction
RNA from cells or tissues was extracted with TRIzol™ (Invitrogen); 2 µg total RNA for each sample was reverse-transcribed using the Oligo dT primer by Superscript™ II (Invitrogen). RT-PCR was performed with AccuPower™ PCR PreMix (Bioneer, Daejeon, Korea). The PCR conditions were as follows: initial melting (94°C, 2 min), 35–40 cycles of amplification (94°C, 20 s/45–65°C, 30 s/72°C, 30 s), final extension (72°C, 3 min). Concentration of each cDNA was normalized using the RT-PCR bands of Gapdh.
Western Blotting
Cell extracts were separated by 10% SDS–PAGE and the proteins were transferred to a PVDF membrane (Millipore, Bedford, MA). The membrane was incubated for 1 h in a block solution [5% nonfat dry milk in Tris-buffered saline containing Tween 20 [TBST]) and washed three times with TBST. After washing, the membrane was incubated with the primary antibody overnight, washed three times with TBST and then incubated with HRP-conjugated secondary antibody for 1 h. Immunoreactive bands were visualized using a chemiluminescence detection system (AbFrontier, Seoul, Korea).
Hemocytometer-Based Proliferation Assay
Cells (1 × 105) were set on 60Φ dish containing normal media without antibiotics. The amount of cells was measured three times at the 1st, 3rd, 5th, and 7th days from the onset.
Wounding Assay
Cells were cultured to confluence. Lines were drawn with a marker on the bottom of the dish. Using a sterile 1,000 µl pipet tip, separate wounds were scratched through the cells moving perpendicular to the line drawn. Cells were rinsed very gently with PBS. Pictures were taken at the day of onset and 48 h after. Media were replaced every day. Measurement was performed to 14–20 samples per every group.
Double Thymidine Block and Facs Analysis
Cells (5 × 104) were set on the 60Φ dishes and grown in normal media for a day. Cells were arrested with 2 mM thymidine (Sigma) for 28 h, released in normal media, and arrested again with 2 mM thymidine for 24 h. After media were changed with normal one, cells were harvested at 0, 4, 8, 10, 12, 14, 16, 20, 24, 28, 32, 36, 40, 44, 48, 52, 56, and 60 h, washed with PBS two times and fixed with 200 µl of PBS and 500 µl of ethanol at −20°C for 1 day. To remove the ethanol, cells were washed with PBS one time and centrifuged at 10,000 rpm for 5 min. Staining was performed with 10 µg of propidium iodide (PI, Sigma) and 10 µg of RNase A (Qiagen Inc, Valencia, CA) in 200 µl of PBS. The samples were incubated at 37°C for 10 min. Analysis was performed with BD FACSCalibur™ (BD Bioscience). The population of G0/G1, G2/M, and S was analyzed with ModFit LT software (Verity Software House Inc., Topsham, ME).
DNA/RNA Quantification Using Pyronin Y and Hoechest 33342
Cells were harvested and incubated in 1 ml culture medium containing 10 µg/ml Hoechst 33342 (Sigma) at 37°C for 45 min. Being added with 5 µl of 100 µg/ml Pyronin Y (Sigma), cells were incubated at 37°C for a further 15 min. Cells were transferred onto ice and analyzed with BD FACSCalibur™ (BD Bioscience).
Data Processing and Statistical Analysis
Numerical data were presented as means ± standard error. Two groups of data were compared using Student's t-test. P-value < 0.05 was considered statistically significant. All data were processed with SAS software (SAS institute Inc., Cary, NC).
RESULTS
Partial Pancreatectomy Up-Regulated the Expression of Glul in the Acinar Cells and the Ductal Cells
Compared to the control group (Fig. 1A-a), the pancreas of the experimental group (Fig. 1A-b) demonstrated prominent inflammation with pancreatic parenchyma destruction, mainly in the acinar component. The islet (arrow heads) and ductal components (arrows) were shown to be relatively retained. GLUL was highly expressed in the partial pancreatectomy group (Fig. 1A-d) relative to the control group (Fig. 1A-c). GLUL expression was prominent in the acinar cells and expression was slightly enhanced in the ductal cells (arrows). Expression in the islet cells (arrow heads) was similar to that of control group. A higher level of GLUL was more detected in the inflammatory region than the intact region (Fig. 1A-e). The Glul gene was shown to be up-regulated in heterogeneous pancreatic tissue in the RT-PCR and Western blotting analysis (Fig. 1B and C).
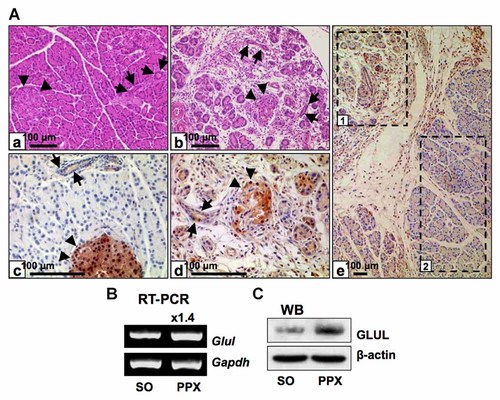
Partial pancreatectomy up-regulated the expression of GLUL in the acinar cells and the ductal cells. A-a: Sham-operated (SO) pancreas, HE staining. A-b: Pancreas remained from partial pancreatectomy (PPX), HE staining. The tissue demonstrates prominent inflammation with pancreatic parenchyma destruction, mainly in the acinar component. The islet (arrow heads) and the ductal components (arrows) are relatively retained. A-c: SO pancreas, immunostaining of GLUL. A-d: PPX pancreas, immunostaining of GLUL. The GLUL expression was prominent in the acinar cells and the expression was slightly enhanced in the ductal cells (arrows). The expression in the islet cells (arrow heads) was similar to that of control group. A-e: PPX pancreas, immunostaining of GLUL. Inflammatory region [1] shows higher expression of GLUL than intact region [2]. B: RT-PCR analysis, heterogeneous pancreas. C: Western blotting, heterogeneous pancreas.
GLUL Increased Cell Growth Rate in PANC-1 Cells and INS-1 Cells
In order to investigate the effect of GLUL on cell growth, either GLUL was overexpressed or down-regulated in both PANC-1 and INS-1 cell lines (Fig. 2A and B). In PANC-1, GLUL-overexpressing cells grew 1.2 times faster than the mock-control cells (Fig. 2C). In INS-1, GLUL-overexpressing cells grew 1.4 times faster than the mock-control cells (Fig. 2E). MSO is an inhibitor of GLUL. When 15 mM MSO was added to the media, PANC-1 showed reduced proliferation but no growth restriction was observed in the INS-1 cells (Fig. 2D and F). The wounding assay for PANC-1 showed similar results with the cell counting assay (Fig. 2G). In this assay, 45% of scratch was recovered for the GLUL-overexpressing cells over 48 h while only 37% of scratch recovered for the mock-control cells over 48 h (Fig. 2H). The enhanced wound recovery of GLUL-overexpressing cells decreased from 45% to 36% when treated with 15 mM MSO (Fig. 2I).
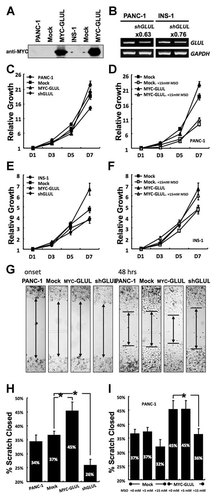
Proliferative effect of GLUL in PANC-1 cells and INS-1 cells. A: GLUL overexpression, Western blotting. B: GLUL knockdown, RT-PCR. C–F: Cell counting assay. Initially, 1 × 105 cells were set on 60Φ dish and counted cells from the next day. Relative growth was calculated based on the cell count number of the 1st day; n = 4. (C. PANC-1, the effect of GLUL. D: PANC-1, the effect of MSO. E. INS-1, the effect of GLUL. F: INS-1, the effect of MSO.) G–I: Wounding assay. Recovery of wound was calculated with ([gap at onset] − gap at 48 h])/(gap at onset). *P < 0.05, n = 14–20. (G. Microscopic image. H: The effect of GLUL. I: The effect of MSO.)
GLUL Overexpression Shifted the Major Population of PANC-1 Cells from G0/G1 Phase to G2/M Phase
Under normal conditions, the cell cycle distribution for PANC-1 and INS-1 cells was as follows: PANC-1, G0/G1-52%, G2/M-27%, S-21%; INS-1, G0/G1-68%, G2/M-9%, S-23% (Fig. 3A). The sub-populations of GLUL-overexpressing cells, GLUL-knockdown cells or MSO-treated cells were compared with sub-population of the mock-control cells (Fig. 3B and C). Only PANC-1 cells overexpressing GLUL showed significantly different sub-population compared with the mock-control cells; G0/G1 and S populations were decreased and G2/M population was increased.
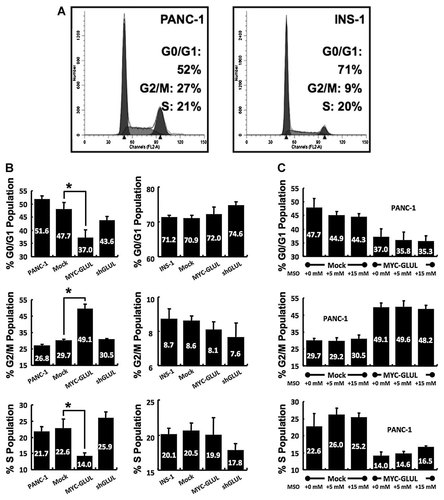
GLUL overexpression shifted the major population of PANC-1 cells from G0/G1 phase to G2/M phase. A: PI staining and FACS analysis; normal PANC-1 and INS-1. B–C: Percentages of sub-populations (G0/G1, G2/M, and S). Each experimental group was compared with mock-control group. *P < 0.05, n = 3. (B: PANC-1 and INS-1, the effect of GLUL. C: PANC-1, the effect of MSO.)
GLUL-Overexpressing Cells Did Not Show Fast Cell Cycle Duration But the Cells Were Resistant to Thymidine Blocking
Double thymidine blocking was used to synchronize the cells into the G1 phase (Fig. 4A). About 78% of the mock-control cells and 46% of GLUL-overexpressing cells were in the G1 and early S phase when treated with 2 mM thymidine (Fig. 4B and C). When the concentration of thymidine was increased to 20 mM, the percent of GLUL-overexpressing cells were enhanced to 55% (Fig. 4C). The synchronized cells showed periodic changes in sub-population through thymidine release. The cycle duration was defined as the mean gap between nearest local maximum points (black arrows in Fig. 4B and D) or local minimum points (gray arrows in Fig. 4B and D). The cycle duration of GLUL-overexpressing cells was 21.7 h, which was similar to the 22.3 h cycle duration of mock cells (Fig. 4E). These cycle durations were shorter than the population doubling time (30–40 h) calculated in the cell counting assay (Fig. 2C).
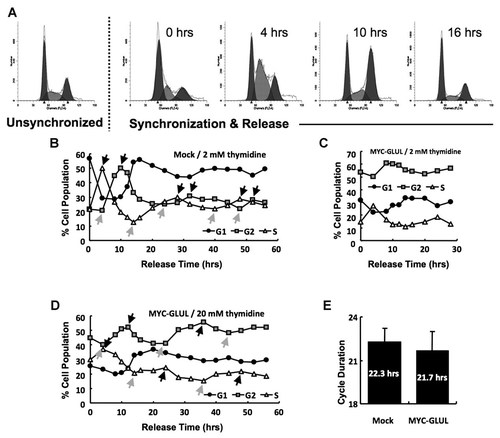
Cycle duration of synchronized cell. A: Double thymidine blocking and release. B: Periodic change of sub-population in synchronized mock cells. Cells were arrested with 2 mM thymidine. C–D: Periodic change of sub-population in synchronized GLUL-overexpressing cells. Cells were arrested with 2 mM (C) or 20 mM (D) thymidine (black arrow—local maximum; gray arrow—local minimum). E: Cell cycle duration of growing cells (calculated by the time gap between nearest local maximums or local minimums in Fig. 4B and D).
GLUL-Overexpressing Cells Have Low G0-Quiescent Population
Pyronin Y staining and FACS analysis was conducted to determine the number of cells in the G0 phase cells; G0-quescent cells have a lower RNA concentration than normal cells (Fig. 5A–C/left lower quadrant). For the negative control, cells were incubated in serum-free media since cells do not grow well under these conditions and most cells remain in the G0/G1 phase. Under these conditions, half of the cells in the G0/G1 population were in the G0-quadrant (Fig. 5A). A majority of the GLUL-overexpressing cells were in the G2/M-phase and their G0 portion was smaller than the G0 portion of mock cells (Fig. 5B and C). Immunocytochemistry was performed using anti-Ki-67. The Ki-67 protein is present during all active phases of the cell cycle but absent in G0-resting cells. While half of the starved cells (serum-free) did not express Ki-67, most mock- and GLUL-overexpressing cells expressed Ki-67 (Fig. 6A–C). Ki-67 was prominently localized to the perinucleolar region in the mock cells; however, it was dispersed at the karyoplasm in a number of GLUL-overexpressing cells. GLUL-overexpressing cells had a slightly higher percentage of Ki-67 positive cells than the mock cells (Supplementary Fig. 1). More GLUL-overexpressing cells were shown to express Ki-67 in the FACS analysis (Fig. 6D).
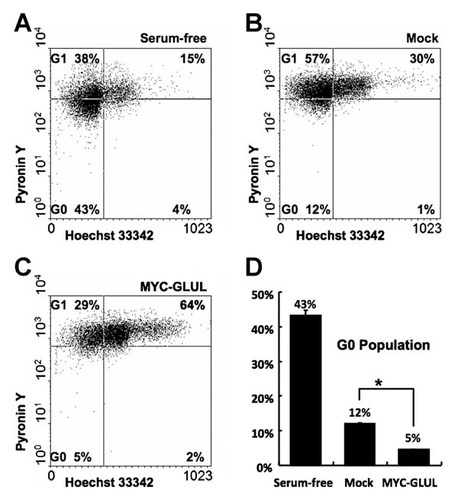
Low G0 population of GLUL-overexpressing cells. A–C: FACS analysis, DNA was stained with Hoechst 33342 and RNA was stained with Pyronin Y (A: Cells incubated in serum-free media for 2 days. B: Mock-control cells. C: GLUL-overexpressing cells. D: The percentage of G0 population).

Up-regulated Ki-67 in GLUL-overexpressing cells. Ki-67 (red) expression in PANC-1 cells. Nuclei—Hoechst 33342 (blue). A: In serum-free media, starved cells have a half of Ki-67 negative count. B–C: Mock (B) and GLUL-overexpressing (C) cells have high Ki-67 positive counts. A number of GLUL-overexpressing cells were stained with Ki-67 in karyoplasm rather than perinucleole. D: FACS analysis of the cells stained with anti-Ki-67.
AKT, ERK1/2, JNK, and P70S6K were Activated in GLUL-Overexpressing PANC-1 Cells
PANC-1 cells were maintained in DMEM with 10% FBS and the media were changed every 2 days. Changing the media highly activated several signals including AKT and ERK1/2 and this effect decreased with time. The effect of fresh media was compared between mock-control cells and GLUL-overexpressing cells 2 h after the media change. GLUL-overexpressing cells contained more phosphorylated AKT, ERK1/2, JNK, and P70S6K, in addition to PCNA and c-Fos, which are involved in cell proliferation (Fig. 7). GLUL appeared to enhance the serum sensitivity of PANC-1 cells. In addition, the expression levels of cyclin A, cyclin B, cyclin D, and RB were equal among the mock cells, GLUL-overexpressing cells, and GLUL-knockdown cells; therefore, GLUL did not induce cell cycle arrest during the G1, S, G2, and M phase.
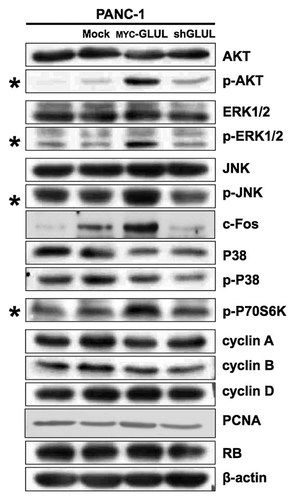
Activation of AKT, ERK1/2, JNK, and P70S6K in GLUL-overexpressing cells. AKT, ERK1/2, JNK, and P70S6K were highly phosphorylated in GLUL-overexpressing PANC-1 cells 2 h after media change. β-actin was used as a loading control for Western blot.
GLUL Weakly Decreasde the Quiescent Population of INS-1 Cells in Glutamine-Free Media
The INS-1 cells grown in glutamine-free media contained a decreased G0/G1 population (Fig. 8A). In the Pyronin Y staining and FACS analysis, GLUL-overexpressing INS-1 cells had a low G0 population compared with mock cells; however, this difference was not significant at the 95% confidence level (P = 0.08, n = 3) (Fig. 8B and C). The enhanced activations of AKT and ERK observed in PANC-1 cells were not detected in INS-1 cells (Fig. 8D). Nevertheless, GLUL-overexpressing INS-1 cells grew 2.4 times faster than mock cells in glutamine-free media (Fig. 8E).

Proliferation of INS-1 cells in glutamine-free media. A: PI staining and FACS analysis. By GLUL overexpression, G0/G1 population was decreased and G2/M population was increased in glutamine-free media. *P < 0.05, n = 3. B–C: Hoechst 33342/Pyronin Y staining and FACS analysis. In glutamine-free media, GLUL-overexpressing cells have smaller G0 population than mock cells. P = 0.08, n = 3. D: Western blot. E: Cell counting assay, n = 4.
DISCUSSION
Glutamine is an amino acid that contains two amine components. Glutamine is converted from glutamate and the balance of these two compounds is important to the regulation of ammonia metabolism and neuroendocrine signaling. GLUL is a crucial enzyme that regulates the conversion between glutamine and glutamate. Since the conversion of glutamine and glutamate is not fast under normal conditions, their distribution depends on intercellular or inter-organ flux [Squires and Brosnan, 1983; Darmaun et al., 1986]. Disease and injury introduce metabolic deficiency in local tissues, and distort the composition of the plasma and tissue. Glutamine in plasma or skeletal muscle was decreased in critically ill patients [Gamrin et al., 1996] and plasma glutamine level was reduced by endotoxin-induced systemic inflammation [Andreasen et al., 2009]. Since inflammation reduced plasma glutamine level, the up-regulation of GLUL prevented metabolic changes [He et al., 2010]. In our study, the up-regulation of GLUL was clearly observed in pancreatic acinar cells which do not regulate GLUL under normal conditions. In acute pancreatitis, glutamine was decreased and glutamate was increased [Roth et al., 1985]. Glutamate and glutamine regulate the response to inflammation. However, the efficiency of this regulation is different from each other. In the case of HSP25 induction, glutamine was a better efficient inducer than glutamate [Phanvijhitsiri et al., 2006]. Glutamine reciprocally down-regulated genes induced by pro-inflammatory cytokines [Brasse-Lagnel et al., 2007] and up-regulated anti-inflammatory genes [Ziegler et al., 2005; Singleton and Wischmeyer, 2006]. Glutamine-deprived cancer cells were shown to be susceptible to complement mediated cell lysis [Ellison et al., 2007].
The activity of GLUL was related to liver cell proliferation and age [Wu, 1984]. Similarly, the amount of GLUL was increased in partial pancreatectomy and overexpression of GLUL regulated PC-12 cell growth and proliferation [Choi et al., 2010]. The ammonia detoxification by GLUL was a critical factor of cell proliferation [Tang et al., 2008]. Amino acid response elements regulated gene expression through ATFs, C/EBPs, and other factors [Brasse-Lagnel et al., 2009]. Glutamine directly participates in protein synthesis, amino acids conversion, nucleotide synthesis, cellular transcription, and ammonia detoxification and the proliferative effect is the collective result of these multi-functions of glutamine [Wasa et al., 1996]. When the protein source of glutamine is limited, cells require the active machinery of amino acid conversion, GLUL. In the pancreas, the expression of GLUL is not high although its mRNA is easily translated to protein [Lund, 1970; Straaten et al., 2006]. The proliferative effect of GLUL was different between PANC-1 and INS-1 cells which resulted from the different characteristics of the two cell lines (the intrinsic doubling time, the endogenous activity of GLUL, and the cell-specific function of glutamine). The doubling time of PANC-1 was 2 days while the doubling time of INS-1 was 3 days. PANC-1 was not highly sensitive to the culture conditions while INS-1 was very sensitive to the culture conditions.
We also examined the effect of GLUL of the cell cycle. PI staining and FACS analysis was performed to address this question. In the assay, only GLUL-overexpressing PANC-1 cells had altered cell sub-populations. Based on the previous proliferation assay, the increased G2/M populations indicate that GLUL might shorten the overall cell cycle regardless of G2/M arrest. Thymidine blocking and release were analyzed to measure the cycle duration. Unexpectedly, the cycle duration of GLUL-overexpressing cells was similar to the cycle duration of mock cells. Nevertheless, this experiment showed that GLUL-overexpressing cells were resistant to thymidine blocking. Regarding thymidine metabolism, GLUL-overexpressing cells appeared to have an advantage in processes that involve nucleotides.
We, then, focused on the high G0/G1 population in mock-control cells to better understand the increased proliferation of GLUL-overexpressing cells. FACS analysis using Pyronin Y staining showed that the G0 population of GLUL was 7% smaller than the G0 population of the mock-control cells. This phenomenon was confirmed by Ki-67 staining. A previous study reported that excess glutamine allowed 3T3-fibroblasts to continue through the cell cycle in the serum-starved state [Zetterberg and Engstrom, 1981]. GLUL-overexpressing cells highly activated AKT, ERK1/2, JNK, and P70S6K. These signals have been shown to be related to the regeneration of the pancreas [Dai et al., 2009; Uzan et al., 2009]. AKT and ERK1/2 regulate various proliferative signals including the cell cycle, protein synthesis, anti-apoptosis, etc. [Maddika et al., 2008]. The mTOR signal up-regulates mRNA translation and ribosome biogenesis when triggered by growth factors, ATP/AMP, and amino acids [DeBerardinis and Cheng, 2010; Silvera et al., 2010]. The mTOR signal and RNA metabolism can be used to discriminate the states of quiescence and proliferation [Bullwinkel et al., 2006; Yanagida, 2009]. GLUL is known to regulate cell quiescence through cellular glutamine levels and this may explain the replication of mature pancreatic cells.
In this study, we showed that GLUL was up-regulated in the pancreas tissue after partial pancreatectomy and may be a critical factor for pancreatic proliferation under stress conditions. Since inflammatory response enhances β-cell regeneration [Ablamunts et al., 2007], GLUL up-regulation might play a role in the balance between inflammation and regeneration. Glutamine affects the expression of genes involved in insulin secretion and cellular integrity [Newsholme et al., 2007] and has been used in total parental nutrition for the management of acute pancreatitis [Ockenga et al., 2002]. Therefore, glutamine appeared to be important to pancreatic physiology. We showed that GLUL could regulate the quiescence of pancreatic cells. By monitoring GLUL expression in the pancreas, it will be possible to develop methods to protect this organ from disease and senescence.
Acknowledgements
We thank Dr. Kimberly Kim at Dana-Farber Cancer Institute, Harvard Medical School in Boston, MA, USA, for her critical comments on the manuscript and Jang-Joon Park and So-Ra Kim for Figure 7. This study was supported by a grant from Korea Health 21 R&D Project, Ministry of Health, Welfare and Family Affaires, Republic of Korea (01-PJ10-PG6-01GN13-0002).




