Molecular characterisation of a serum-responsive, DAF-12-like nuclear hormone receptor of the fox-tapeworm Echinococcus multilocularis
Abstract
As the primary mediators of lipophilic and steroid hormone signalling, the family of nuclear receptors (NRs) plays a central role in the regulation of metazoan development. Lipophilic hormones are also thought to be important players in the molecular interaction between larval cestodes and their hosts but no member of the NR family has yet been characterised in this group of parasites. In this work, we provide for the first time evidence for the presence of NRs in cestodes of the genus Echinococcus. By bioinformatic analyses, we identified a set of 17 NRs in the genomes of E. multilocularis and E. granulosus which broadly overlapped with the set of NRs that is expressed by schistosomes, but also contained several members that are unique to cestodes. One of these receptors, EmNHR1, displayed structural homologies to the DAF-12/HR-96 subfamily of NRs that regulates cholesterol homeostasis and longevity in metazoans. By RT-PCR analyses, we demonstrate that the EmNHR1 encoding gene is expressed in all Echinococcus larval stages that are involved in the infection of the intermediate host. By yeast two-hybrid analyses, we further demonstrate cross-communication between EmNHR1 and TGF-β signalling pathways in Echinococcus and that mammalian serum contains a ligand that induces homodimerisation of the EmNHR1 ligand-binding domain. EmNHR1 could thus play an important role in hormonal host–parasite cross-communication mechanisms during an infection. On the basis of our results, further investigations into the role of NR signalling in cestode development and host–parasite interaction will be greatly facilitated. J. Cell. Biochem. 112: 1630–1642, 2011. © 2011 Wiley-Liss, Inc.
The larval stage of the fox-tapeworm Echinococcus multilocularis is the causative agent of alveolar echinococcosis (AE), one of the most serious and lethal parasitoses in humans [Kern, 2010]. The E. multilocularis life cycle comprises an adult stage that dwells in the intestine of the definitive host (foxes or dogs) and three successive larval stages (oncosphere, metacestode and protoscolex) that are involved in the infection of the intermediate host (rodents, occasionally humans). Infiltrative, tumour-like growth of metacestode tissue within the intermediate host's liver, accompanied by metastasis formation in secondary organs during long-term infections is a hallmark of AE [Brehm et al., 2006; Kern, 2010]. Due to its accessibility to in vitro cultivation, E. multilocularis has during recent years been established as a laboratory model system for studying molecular interactions between mammalian hosts and larval cestodes, of which the dog-tapeworm E. granulosus and the pork-tapeworm Taenia solium are of world-wide medical importance [Brehm et al., 2006; Brehm, 2010a]. The respective methodology includes in vitro cultivation systems by which early parasite development from totipotent stem cells (neoblasts) of the oncosphere towards the metacestode [Spiliotis et al., 2008, 2010], as well as late developmental phases from mature metacestode vesicles towards the production of protoscoleces [Hemphill et al., 2002; Spiliotis et al., 2004; Brehm and Spiliotis, 2008; Spiliotis and Brehm, 2009] can be reconstituted in vitro. Using these cultivation systems, evidence has been obtained that parasite development is governed by growth factors that are secreted by co-cultivated host feeder cells and/or are present in host serum [Hemphill et al., 2002; Brehm, 2010a,b]. Furthermore, we could molecularly characterise a number of evolutionarily conserved Echinococcus signalling systems of the insulin-, the epidermal growth factor (EGF)- and the transforming growth factor-β (TGF-β)/bone morphogenetic protein (BMP) pathways [Brehm, 2010a,b] and demonstrated that host derived insulin, BMP2 and EGF can interact with the respective Echinococcus surface kinases and/or stimulate the parasite's mitogen-activated protein kinase (MAPK) pathway [Brehm et al., 2006; Gelmedin et al., 2010; Brehm, 2010b]. Since inhibition of the Echinococcus MAPK pathways drastically affects parasite development and proliferation [Gelmedin et al., 2008, 2010], stimulatory host cytokines of the EGF- and insulin families seem to play an important role in host–parasite interaction during AE [Brehm, 2010b].
Although substantial work has already been carried out concerning the influence of peptide hormones and cytokines on Echinococcus development, questions concerning related effects of steroid and lipophilic host hormones have so far not been addressed, nor have corresponding receptor sequences been described in any cestode. Lipophilic hormone signalling crucially involves so-called nuclear receptors (NRs), DNA-binding transcription factors that are essential in animal development, differentiation and homeostasis [Aranda and Pascual, 2001; Robinson-Rechavi et al., 2003; Novac and Heinzel, 2004; Wu and LoVerde, 2010]. NRs form a superfamily of phylogenetically related proteins and share a universal modular structure consisting of five to six domains, designated A to F, from the N-terminus to the C-terminus. The N-terminal regulatory domain (region A/B) contains the activation function 1 (AF1), the activity of which is independent of ligand binding. The DNA-binding domain (DBD, region C) is highly conserved among NRs and contains two zinc finger motifs of which the first (containing the P-box) mediates binding to specific hormone responsive elements (HREs) in the promoter regions of target genes, whereas the second (D-box) is involved in receptor dimerisation. The ligand-binding domain (LBD, region D) is only moderately conserved among NRs and contains the activation function 2 (AF2), the activity of which is dependent on the presence of bound ligand [Novac and Heinzel, 2004; Wu and LoVerde, 2010]. Apart from interacting with the cognate ligand, the LBD is also involved in heteromeric interactions with co-activators and co-repressors that modulate NR activities [Aranda and Pascual, 2001].
The number of NRs that are expressed by different vertebrate and invertebrate species varies considerably. In the human genome, 48 different NR-encoding genes were identified [Robinson-Rechavi et al., 2003] whereas 18 are encoded on the genome of Drosophila melanogaster [King-Jones and Thummes, 2005] and more than 270 by the model nematode Caenorhabditis elegans [Sluder et al., 1999]. In platyhelminths, NRs have so far only been reported for the parasitic trematode species Schistosoma mansoni and S. japonicum, the genomes of which have been fully sequenced [Webster et al., 2010]. In S. mansoni, 21 NRs were identified and characterised concerning their expression patterns throughout the life cycle, the formation of homo- and heterodimers, and their DNA binding capabilities [reviewed by Wu and LoVerde, 2010]. Interestingly, these studies also revealed that S. mansoni expresses three unusual NRs with two DBDs and one LBD (the so-called 2DBD-NRs) that were later also identified in other lophotrochozoans such as planarians and molluscs [Wu et al., 2006]. S. japonicum expresses virtually the same set of NRs as S. mansoni with the exception that it contains two additional members of the 2DBD-NR family [Wu and LoVerde, 2010]. Although several reports have been published that the cestode Taenia crassiceps is capable of producing NR ligands such as steroid hormones [Jimenez et al., 2006; Fernandez-Presas et al., 2008] and displays certain in vitro responses to host-derived steroids [Escobedo et al., 2004], no structural evidence for the expression of NRs in T. crassiceps or in any other cestode has been reported so far. In this work, we provide evidence for the presence of such receptors in a cestode for the first time. We demonstrate that E. multilocularis expresses an NR, EmNHR1, with structural homologies to DAF-12 and NR-96 that control lipid homeostasis and longevity in C. elegans and D. melanogaster, respectively. We provide evidence that EmNHR1, like other members of the DAF-12/NR-96 family, cross-interacts with TGF-β-signalling in E. multilocularis and that a ligand for EmNHR1 is present in host serum. Furthermore, we present detailed genomic and bioinformatic analyses on the set of NRs that is present in the E. multilocularis genome.
MATERIALS AND METHODS
Organisms and Culture Methods
Parasite material of the E. multilocularis isolate J31 [Gelmedin et al., 2010] were propagated and constantly kept in Mongolian jirds (Meriones unguiculatus) as described previously [Spiliotis and Brehm, 2009]. In vitro cultivation of metacestode vesicles and primary cells was performed essentially as previously described [Spiliotis et al., 2004, 2008; Spiliotis and Brehm, 2009]. Protoscoleces were isolated from in vivo cultivated parasite material according to a previously established protocol [Brehm et al., 2003] and were activated by pepsin/low pH treatment as described by Gelmedin et al. [2010].
Nucleic Acid Isolation, Cloning and Sequencing
Chromosomal DNA was isolated from parasite material after cultivation in M. unguiculatus as outlined previously [Brehm et al., 2000]. RNA isolation from in vitro cultivated axenic metacestode vesicles and protoscoleces was performed using TRIZOL®-Reagent (cat.-no. 15596-018; Invitrogen, Karlsruhe, Germany) essentially as described [Gelmedin et al., 2010]. For RNA isolation of axenically cultivated Echinococcus primary cells, a modified protocol was applied using 500 µl of TRIZOL®-Reagent and 100 µl of chloroform. For reverse transcription, 2 µg total RNA was used and cDNA synthesis was performed as previously described using oligonucleotide CD3-RT [Brehm et al., 2003]. PCR products were cloned using the TOPO-TA cloning kit (Invitrogen) or the PCR Cloning Kit (QIAGEN, Hilden, Germany) and sequenced employing an ABI prism 377 DNA sequencer (Perkin-Elmer, Rodgau-Juegesheim, Germany).
Cloning and Characterisation of the Emnhr1 cDNA and Genomic Locus
Clone Z3-214 was identified after random sequencing of the inserts of a previously established cDNA library of trans-spliced transcripts [Brehm et al., 2000]. The clone was fully sequenced using vector specific primers (M13 forward: 5′-GTA AAA CGA CGG CCA GT-3′; M13 reverse: 5′-CAG GAA ACA GCT ATG ACC-3′) and insert specific primers (NHR1-C3: 5′-CTG CAG TAC TTG GCG TGC TC-3′; NHR1-C4: 5′-GGA CTC CAT GCC TCC AAA CG-3′; NHR1-C6: 5′-GAC TTG CGC ATT CCC ACA GC-3′). The genomic locus of Emnhr1 was characterised using different PCR primers directed against the cDNA sequence of Emnhr1 after amplification in form of overlapping fragments from chromosomal E. multilocularis DNA. The Emnhr1 cDNA sequence has been deposited in the GenBank™ database and is available under the accession number FN869039.
RT-PCR Analyses
Equal amounts of cDNA were subjected to RT-PCR analysis using primers EmNHR1-LHQ-dw (5′-CTT CAT CAG GAT CGC TTA CCT AC-3′) and EmNHR1-SLV-up (5′-TCA CCA AAC TAC TTA TAG TAA TAT TTG AG-3′) for Emnhr1 and, as a control, Em10-15 (5′-AAT AAG GTC AGG GTG ACT AC-3′) and Em10-16 (5′-TTG CTG GTA ATC AGT CGA TC-3′) directed against the constitutively expressed gene Emelp [Brehm et al., 2003] employing a protocol of 35 cycles of 30 s at 94°C, 30 s at 57°C and 50 s at 72°C. The resulting PCR products were separated on a 1% agarose gel and stained with ethidium bromide. The expected PCR product sizes from cDNA were 582 bp (Emnhr1) and 450 bp (Emelp).
Yeast Two-Hybrid Analyses
To study protein–protein interactions in a yeast two-hybrid approach the MATCHMAKER™ system (Clontech) was used as previously described [Zavala-Gongora et al., 2003, 2008]. Briefly, translational fusions of the proteins or protein domains of interest with the Gal4 DBD (pGBKT7; BD) or the Gal4-activation domain (pGADT7; AD) were generated. Fusion constructs were co-transformed into yeast strain AH109 and positive transformants were selected in leucine/tryptophan-deficient minimal medium. Proper expression of the fusion proteins was verified by Western blot analyses using antibodies against the Gal4-AD (anti-HA; SantaCruz Technologies) and Gal4-BD (anti-cmyc; SantaCruz Technologies). Interaction trap analysis of double transformants was performed according to the MATCHMAKER™ instruction manual using selective agar plates lacking leucine, tryptophan (as plasmid selection markers) and either histidine in the presence of 7.5 mM 3-amino-triazole (3-AT) for medium stringency or histidine plus adenine for high stringency selection. Colony formation was assessed after 3 days of incubation at 30°C. For analysis in the liquid culture β-galactosidase assay, constructs were co-transformed into yeast strain Y187. The assay was performed essentially according to the MATCHMAKER™ manual. Briefly, over-night cultures of co-transformants were grown for 5 h in leucine/tryptophan deficient medium to comparable density (OD600 nm). Yeast cells were then harvested and washed in Z buffer (16.1 g/L Na2HPO4·7H2O; 5.50 g/L NaH2PO4·H2O; 0.75 g/L KCl; 0.246 g/L MgSO4·7H2O; pH 7.0). After three freeze-and-thaw cycles in liquid nitrogen, Z buffer/0.27% (v/v) β-mercaptoethanol and 0.67 mg/ml o-nitrophenyl β-D-galactopyranoside were added to the lysates. The reaction was stopped with 1 M Na2CO3 once the yellow colour had developed.
The AD and BD constructs for full-length EmNHR1, the DBD, and the complete as well as a C-terminally truncated form of the LBD have been obtained by directional cloning. First, full-length Emnhr1 was cloned into plasmid pDrive (Qiagen PCR cloning kit) after amplification from metacestode cDNA using primers NHR1_H95_ATG_NdeI_dw (5′-AAT CCT CAT ATG ACG TCC CAA AAG CAA G-3′) and NHR1_H95_TAA_up (5′-TTA GGA GGA ATT AGT CAT GTT AG-3′) yielding plasmid pDrive-NHR1. The EmNHR1 DBD was amplified from pDrive-NHR1 using primers NHR1_H95_ATG_NdeI_dw (see above) and NHR1_H95_DBD_Y2H_XmaI_up (5′-CCG TGG CCC GGG GAG ATG CTG AGA TCA AAG G-3′). For LBD constructs, the following primers were chosen: EmNHR1-LHQ-NcoI-fwd (5′-TGC CCA TGG ATC TTC ATC AGG ATC GCT TAC C-3′), EmNHR1-FIE-XmaI-rev (5′-CAC CCC GGG CAC TCG ATA AAC ATA TGT ACC-3′) and EmNHR1-ALT-XmaI-rev (5′-TAG CCC GGG GTC AGA GCC TGT AAC TCC G-3′; C-terminally truncated). Insertion into pGADT7 and pGBKT7 was achieved by Nde I/Xma I restriction digest for EmNHR1-DBD and by Nco I/Xma I restriction digest for both LBD constructs. Full-length Emnhr1 was introduced into pGADT7 via Nde I/Sac I and into pGBKT7 via Nde I/Sal I restriction digestion. The cloned inserts were fully sequenced to ensure that no mutations had been introduced due to incorrect PCR amplification. Yeast two-hybrid fusion plasmids for EmSmadA, EmSmadB, EmSmadC, EmSmadD and EmSkip have previously been described [Zavala-Gongora et al., 2003, 2008; Gelmedin et al., 2005].
In Silico and Statistical Analyses
Amino acid comparisons were performed using the basic local alignment search tool (BLAST) on the nr-aa database collection available under http://blast.genome.jp. Pileups and CLUSTAL W alignments were constructed employing the software BioEdit Sequence Alignment Editor (version 7.0.0) using the BLOSUM62 matrix. Domain predictions were performed using the simple modular architecture research tool (SMART) available under http://smart.emblheidelberg.de. Genomic analyses and BLAST searches against the current assembly version of the E. multilocularis genome were done using the respective resources of the Sanger Institute (Hinxton, UK) available under http://www.sanger.ac.uk/cgi-bin/blast/submitblast/Echinococcus. The identification of putative E. multilocularis NRs was carried out in two steps: an iterative tBLASTN was performed against the E. multilocularis genome assembly with the deduced amino acid sequence of the EmNHR1 DBD using the WinBlast software (v.0.2.0) with an E-value cut-off at 1.0. With all DBD sequences obtained by this approach, and DBD sequences of the complete set of S. mansoni NRs [Wu and LoVerde, 2010], additional BLAST searches against the available genome information were carried out to completely identify the parasite's NR set. After confirming the characteristic zinc-finger structure of the identified DBDs by SMART analysis, BLASTX analyses at http://blast.ncbi.nlm.nih.gov/Blast.cgi were performed for all identified genes. To this end, nucleotide sequence regions obtained from the tBLASTN analysis were retrieved using boundaries of 2000 nt downstream and 5000 nt upstream of the respective sequence. The data collection of putative E. multilocularis NR includes the best hit (lowest E-value; highest bit score) for each BLASTX search with its respective tBLASTN sequence. The respective sequences were aligned using CLUSTAL W.
For phylogenetic tree reconstruction, sequences were aligned with CLUSTAL W software and then transferred to MEGA 4.0 using the NeighbourJoining complete deletion method with a bootstrap value of 1000. For statistical analyses a two-tailed, unpaired Student's t-test was performed. Error bars represent standard error of the mean. Differences were considered significant for P-values below 0.05 (indicated by *).
Ethics Statement
All experiments were carried out in accordance to the European and German regulations on the protection of animals (Tierschutzgesetz, Section 6). Ethical approval of the study was obtained by the local ethics committee of the government of Lower Franconia (621-2531.01-2/05).
RESULTS
Isolation and Characterisation of the Emnhr1 CDNA and Genomic Locus
In previous investigations on the mechanism of mRNA trans-splicing in E. multilocularis [Brehm et al., 2000, 2003], cDNA libraries for trans-spliced transcripts from in vitro-cultivated metacestode vesicles had been produced and a number of randomly chosen clones were sequenced. In these analyses, one clone (Z3-214) was identified, the recombinant insert of which coded for a protein with considerable homologies to NRs (Fig. 1). Since this was the first member of this protein family to be described in the parasite, the respective gene was designated Emnhr1 (for E. multilocularis nuclear hormone receptor 1), coding for the protein EmNHR1. The Z3-214 insert was fully sequenced and contained one single reading frame encoding a protein of 651 aa and a theoretical molecular mass of 72 kDa. A putative polyadenylation signal (AATAAA) was present 17 nt upstream of the poly(A) tail.
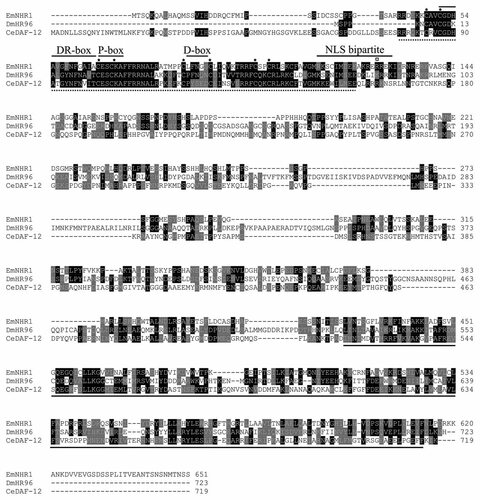
Structural characteristics and amino acid sequence homologies of EmNHR1. In a CLUSTAL W alignment the sequences of E. multilocularis EmNHR1 (this work), D. melanogaster HR-96 (DmHR96; NP_524493) and C. elegans DAF-12 (CeDAF-12; NP_001041239.1) are shown. Amino acid residues matching the consensus are printed in white on black background and residues with similar function are marked white on grey background. The complete LBD domain is indicated by a grey line below the alignment. Structural motifs of the DBD such as the P-, D-, DR-box and NLS bipartite are indicated above the alignment. The conserved Cys residues of the ZnF_C4 motif and an Arg residue for high-affinity DNA binding [Shostak et al., 2004] are marked by asterisks and an open square, respectively.
Homology analyses using the SMART software package revealed the presence of a typical C4 zinc finger (ZnF_C4) DBD in EmNHR1 between residues 22 and 93 as well as a hormone receptor LBD between residues 406 and 567 (Fig. 1). As expected, BLAST analyses in nr-aa and SWISSPROT databases revealed only low overall sequence homologies of EmNHR1 to other proteins. However, significant similarities were detected for the EmNHR1 DBD and the LBD. The EmNHR1 DBD was 57% identical and 68% similar to the DBD of C. elegans DAF-12, which is a homolog of mammalian Liver X receptors, the Pregnane X receptor and vitamin D receptor [Mooijaart et al., 2005]. Regarding the P-box sequence, which is in the N-terminal zinc finger and important for DNA-binding specificity, EmNHR1 can be placed into the invertebrate-specific CESCKA receptor subfamily which also includes the three C. elegans NRs DAF-12, NHR-8 and NHR-48 [Sluder et al., 1999] as well as D. melanogaster HR-96 [Fisk and Thummel, 1995]. Including the P-box sequence, DAF-12, NHR-8, NHR-48 and HR-96 contain 13 contiguous conserved residues (TCESCKAFFRRNA) that form the DNA-recognition α helix [Shostak et al., 2004]. EmNHR1 shares 12 amino acids with this sequence (CESCKAFFRRNA), suggesting that it recognises similar HREs as the nematode and insect orthologs. In addition to the four highly conserved Cys residues that form the zinc finger, EmNHR1 also contains a conserved Arg residue at position 127 that is essential for high-affinity DNA binding [Shostak et al., 2004] and a bipartite nuclear localisation sequence (Fig. 1). Apart from considerable homologies within the DBD domain to members of the DAF-12/HR-96 family of NRs, EmNHR1 also displayed clear similarities to this protein family within the LBD (Fig. 1). Highest homologies were detected in this region to an NHR-48 ortholog of the nematode Brugia malayi (31% identical/51% similar residues), to D. melanogaster HR-96 (28/48) and C. elegans DAF-12 (28/50). Homologies of 27% identical and 43% similar residues were furthermore detected between EmNHR1 and mammalian Pregnane X- and vitamin D receptors. Taken together, these analyses showed that EmNHR1 is a typical NR and is most closely related to the invertebrate DAF-12/HR-96 family that controls cholesterol homeostasis and longevity in insects and nematodes [Mooijaart et al., 2005; Bujold et al., 2010].
The genomic Emnhr1 locus was PCR amplified from E. multilocularis chromosomal DNA and cloned in form of several overlapping fragments. The locus spans a region of 6.4 kb and, within the coding region, 7 introns (between 96 and 2383 bp) with canonical GT-AG splice sites were identified that separated 8 exons of 49–428 bp (Fig. 2). Interestingly, one intron was present in the DBD encoding region at exactly the same position as an intron in the C. elegans DAF-12 gene [Antebi et al., 2000] (Fig. 2). Furthermore, like in the C. elegans DAF-12 gene, the genomic region encoding the EmNHR1 LBD contained two introns of which one was located at a similar position as in C. elegans (Fig. 2). Taken together, these analyses indicated that the E. multilocularis Emnhr1 gene had a recent ancestor with the C. elegans DAF-12 gene.
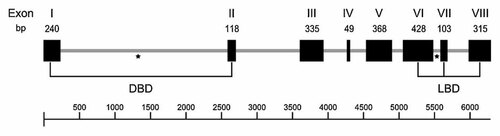
Chromosomal organisation of the E. multilocularis nhr1 locus. The chromosomal region as determined in this study is depicted as a grey line. The bar below represents the scale bar in bp. Exons (I–VIII) are shown as black boxes with the size of each exon (in bp) given above. The locations of coding regions for DBD and the LBD are indicated by black lines. Two introns that are present at identical positions in Emnhr1 and the C. elegans DAF-12 gene are marked by an asterisk.
The E. multilocularis genome is currently being sequenced [Brehm, 2010a] and genome assembly is in a very advanced stage (available at: http://www.sanger.ac.uk/resources/downloads/helminths/echinococcus-multilocularis.html). In order to verify the Emnhr1 sequences we had obtained through PCR amplification and to characterise the genomic context, we performed BLAST analyses on the available contig information. We successfully identified Emnhr1 on contig no. 3395 (scaffold 62) and obtained a sequence that was identical to that which we had identified through direct sequencing of the locus. As expected, we also identified a spliced acceptor consensus sequence directly upstream of the coding region exactly at the position where the cDNA contained the E. multilocularis spliced leader sequence [Brehm et al., 2000]. Immediately adjacent to the Emnhr1 locus, we identified orthologs of the S. japonicum ‘zinc finger protein RTS2’ (GenBank accession number FN314476)-encoding gene (downstream) and a gene encoding subunit 5 of the eukaryotic transcription factor IIIC (upstream). In addition to assembled contigs for E. multilocularis, extensive sequence information for the closely related dog-tapeworm E. granulosus is available at the Wellcome Trust Sanger Institute platform and in respective BLAST analyses we could identify a locus, Egnhr1, encoding a protein with 98% amino acid sequence identity to EmNHR1. This indicated that the nhr1 gene is highly conserved in the genus Echinococcus.
In Silico Identification of Other Putative E. multilocularis NRs
To identify the entire set of NRs that is expressed by E. multilocularis, extensive BLAST analyses on the available genome information were carried out. The hallmark of an NR is the ZnF_C4 zinc finger motif [Wu and LoVerde, 2010] and we first used this motif of EmNHR1 as well as the respective motifs of known NRs from S. mansoni [Wu and LoVerde, 2010] in tBLASTN searches to identify related sequences in the genome. On the basis of the identified ZnF_C4 motifs, iterative BLAST searches were carried out to determine the entire set of NRs in E. multilocularis. We then analysed the genomic regions that coded for ZnF_C4 motifs for the presence of LBD encoding regions (between 2 kb downstream and 5 kb upstream of the ZnF_C4 region; see also Supplementary Table I). By these analyses, we identified a total set of 17 NRs encoded on the E. multilocularis genome (Fig. 3; Table I). A phylogenetic tree in which the DBDs of the E. multilocularis NRs are compared to those of S. mansoni is depicted in Figure 3 and shows that most of the Echinococcus NRs do have close orthologs in the trematode. Significant similarities were also observed for many Echinococcus and schistosome NRs in the LBD (Table I). A new, invertebrate-specific family of NRs with two ZnF_C4 DBDs and one LBD (2DBD-NRs) has previously been identified in S. mansoni [Wu et al., 2006], which expresses three isoforms of this family. As in the trematode, three isoforms of this family are also encoded on the E. multilocularis genome (contigs 1779, 1872 and 2681) and display significant homologies to the three schistosome 2DBD-NRs in both the DBDs and the LBDs (Fig. 3; Table I). Additional Echinococcus NRs with clear homologies to schistosome orthologs within the DBD and the LBD are encoded by genes present on contigs 1644 (SmTR4), 1680 (SmTRβ), 1702 (SmHNF4), 2122 (SmE78), 3309 (SmHR96α), 3400 (SmFTZ-F1α), 3425 (SmNR4A5) and 3442 (SmFTZ-F1) (Fig. 3; Table I; nomenclature of the schistosome NRs according to Wu and LoVerde [2010]). Only for one putative Echinococcus NR, encoded on contig 2891, no LBD could be identified in our analyses. For another putative Echinococcus NR, encoded on contig 1798, clear homologies to SmTLL were observed in the DBD but not in the LBD (Fig. 3; Table I) and, interestingly, one additional NR, encoded on contig 3440, displayed homologies within the LBD to SmRXR2 but harboured an untypical P-box (CDSCRA) that is not present in any of the schistosome receptors. True orthologs for both schistosome RXR receptors as well as SmDSF, SmPNR or SmRAR-like were not found in the E. multilocularis genome which, unlike S. mansoni, contained an additional member of the CESCKA family, EmNHR1 (Fig. 3; Table I). Interestingly, the genomes of both parasites contain NRs that can be classified as HR96-like molecules (SmHR96α and an Echinococcus ortholog encoded on contig 3309) on the basis of a CESCKA P-box and certain homologies to Drosophila HR-96 and C. elegans Daf-12 within the LBD (12–15% identical, 26–31% similar residues; Fig. 4). Among each other, these NRs display considerable LBD homologies (60% identical, 76% similar residues) but both are only distantly related to EmNHR1 in the LBD (15% identical, 31% similar residues; Fig. 4). Furthermore, the LBDs of these NRs contain two insert regions when compared to the LBDs of DAF-12, HR-96 and the liver X receptor which are not present in the LBD of EmNHR1 (Fig. 4). Hence, although EmNHR1, the Echinococcus NR encoded on contig 3309, and SmHR96α are all members of the HR-96/Daf-12 CESCKA family, EmNHR1 is in so far unique as it displays the highest homologies to the nematode and insect receptors in the LBD and that it does not have a clear ortholog in schistosomes. When we carried out similar analyses on the E. granulosus genome sequence, we could identify the same set of NRs as in E. multilocularis (data not shown). Taken together, these analyses showed that cestodes of the genus Echinococcus contain less NR encoding genes than schistosomes, which is mostly due to the absence of true RXR orthologs and the presence of only one member of group 2E NRs which, in schistosomes, is made up of the receptors SmTLL, SmPNR and SmDSF [Wu and LoVerde, 2010]. On the other hand, Echinococcus contains more receptors with DBD and LBD similarities to the HR-96 group, including EmNHR1.
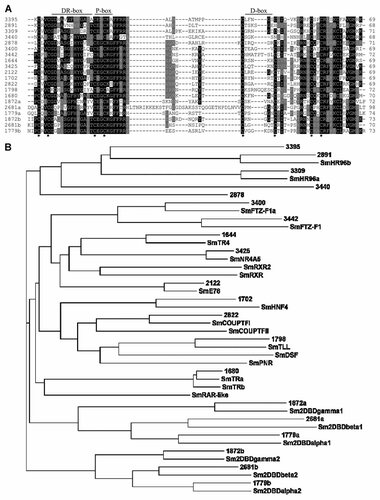
In silico identification of putative NHR in E. multilocularis. A: Amino acid sequence comparison of the DBDs of 17 E. multilocularis NRs identified in this study. The contig numbers of the current E. multilocularis genome assembly that encode the NRs are indicated to the left. In the case of members of the 2DBD family, the ZnF_C4 motifs are marked by a (most N-terminal motif) and b (most C-terminal motif). Residues that match the consensus are printed in white on black background, residues with similar function are marked in grey. Conserved DBD structures such as the P-, DR- and D-boxes are indicated above the alignment. B: Phylogenetic tree depicting DBD homologies between the 17 E. multilocularis NRs and 21 NRs identified in S. mansoni [Wu and LoVerde, 2010]. The E. multilocularis NRs are indicated by the contig number of the present genome assembly. The nomenclature of the S. mansoni NRs was chosen according to Wu and LoVerde [2010].
| Contiga | tBLASTN, positionb ZnF_C4 | SMART, E-value | tBLASTN, positionb LBD | SMART, E-value | Homologiesc |
|---|---|---|---|---|---|
| 1644 | 481637–481467 | 2.6E−30 | 479201–476960 | 3.0E−04 | 60/80% SmTR4 |
| 30/44% NRsubfam 2 Taebiopygia guttata (rs:XP_002199588) | |||||
| 1680 | 186953–187156 | 2.8E−27 | 188485–188779 | 6.6E−02 | 33/44% SmTRb |
| 25/43% ecdysone receptor Liocheles austr. (rs:NP_001037331) | |||||
| 1702 | 102326–102150 | 2.6E−31 | 101830–100228 | 3.3E−24 | 39/58% SmHNF4 |
| 31/50% hep nucl factor 4 Tribolium castaneum (rs:XP_968613) | |||||
| 1779 | 88954–86636 | 1.6E−22 | 81852–79231 | 3.3E−01 | 55/72% Sm2DBDγ |
| 85722–83572 | 2.4E−29 | 31/50% ecdysone receptor Bombyx mori (rs:NP_001037331) | |||
| 1798 | 6287–6364 | 6.1E−26 | 12365–16089 | 1.2E−03 | No homology to S. mansoni |
| 25/43% NR subfam. 6 Danio rerio (sp:GCNFB_DANRE) | |||||
| 1872 | 361173–363091 | 9.5E−14 | 369302–370736 | 3.0E−17 | 34/56% Sm2DBDγ |
| 365772–366071 | 1.4E−29 | 28/46% retinoic acid rec. Lymnaea stagnalis (tr:D5LIR6_LYMST) | |||
| 2122 | 200727–203164 | 6.6E−32 | 211021–213025 | 1.7E−19 | 50/64% SmE78 |
| 36/50% ecdysone-ind. Prot. 78c D. melanogaster (rs:NP_996135) | |||||
| 2681 | 142976–143859 | 7.8E−10 | 151150–153008 | 3.4E−06 | 52/73% Sm2DBDβ |
| 144838–145255 | 5.9E−33 | 26/50% oxysterol receptor LXRb Homo sapiens (tr:B4DNM6_HUMAN) | |||
| 2822 | 79036–79239 | 2.3E−26 | 80631–81051 | 1.0E−12 | No homology to S. mansoni |
| 32/49% NHR-69 C. elegans (sp:NHR_69_CAEEL) | |||||
| 2878 | 337761–335686 | 8.0E−26 | 326253–323919 | 5.3E−08 | No homology to S. mansoni |
| 25/48% nuclear receptor NR1D2 H. sapiens (tr:B4DDP1_HUMAN) | |||||
| 2891 | 140405–144600 | 1.3E−11 | — | — | No LBD |
| 3309 | 400163–399395 | 2.63E−21 | 384020–380850 | 3.6E−02 | 62/77% SmHR96α |
| 26/48% nuclear rec. Strongyloides stercoralis (tr:Q9X2J5_9BILA) | |||||
| 3395 NHR1 | 9827–9726 | 1.91E−18 | 4707–3832 | 3.5E−12 | 17/31% SmHR96α |
| 28/48% NHR96 D. melanogaster (sp:HR96_DROME) | |||||
| 3400 | 259948–259748 | 1.29E−24 | 257206–250996 | 1.3E−11 | 44/70% SmFTZ-F1-α |
| 29/46% NR5A5 Danio rerio (tr:B3KN2_DANRE) | |||||
| 3425 | 163477–164189 | 5.95E−29 | 166248–167938 | 1.5E−02 | 37/66% SmNR4A5 |
| 31/44% retinoic acid rec. Lepisosteus platyrhincus (tr:B7X6R1_9ACTI) | |||||
| 3440 | 155860–156036 | 6.11E−08 | 156852–158018 | 1.6E−08 | 22/40% SmRXR2 |
| 31/50% RXRγ Xenopus laevis (sp:RXRG_XENLA) | |||||
| 3442 | 72501–72713 | 4.51E−20 | 73591–74555 | 1.1E−23 | 64/79% SmFTZ-F1 |
| 40/60% FTZ-F1β D. melanogaster (sp:FTF1B_DROME) |
- a Contig number of the current E. multilocularis genome assembly.
- b Position of the DBD and LBD on the respective contig. In 2DBD members, both ZnF_C4 motifs are indicated.
- c Highest LBD homologies (% identical/% similar residues) to NRs of S. mansoni (above) or to proteins present in the nr-aa database (http://www.genome.jp).

Homologies between EmNHR1 and different members of the DAF-12/HR-96 family of NRs. Depicted is an amino acid sequence alignment between the LBDs of EmNHR1, D. melanogaster HR-96 (DmHR96; GenBank accession no.: Q24143), C. elegans DAF-12 (CeDAF12; AF136238), the human liver X receptor α (HsLXR; P55055), S. mansoni SmHR96α (SmCAR; AY663841) and the Echinococcus SmHR96a-like NR encoded on contig 3309 (Em3309). Residues matching the consensus are printed in white on black background, residues with similar function are marked in grey.
Expression of Emnhr1 and Other NRs in Echinococcus Larval Stages
In Drosophila and C. elegans, HR-96 and DAF-12 interact with components of cholesterol metabolism [Gerisch et al., 2007; Horner et al., 2009] and are most probably acting as sentinels for low cholesterol concentrations [Mooijaart et al., 2005; Bujold et al., 2010]. Since E. multilocularis is not able to synthesise cholesterol de novo and has to take it up from the host [Bernthaler et al., 2009], we were particularly interested in the functions of EmNHR1 and analysed this receptor further. We first investigated the expression of Emnhr1 in Echinococcus larval stages by semi-quantitative RT-PCR. As shown in Figure 5, Emnhr1 transcripts were clearly detected in regenerating primary cells (corresponding to the oncosphere–metacestode transition stage [Spiliotis et al., 2008]), in metacestode vesicles and in protoscoleces prior to (dormant) and after (activated) treatment with pepsin/low pH (mimicking the transition into the definitive host), although higher expression was observed in the metacestode and the non-activated protoscolex. In parallel to genome sequencing, next generation transcriptome profiling is currently carried out at the Sanger Institute on all the larval stages mentioned above using a standard Illumina platform. When analysing the respective dataset, we also observed Emnhr1 expression in all larvae with about 3–4 times higher expression in metacestode vesicles and non-activated protoscoleces (data not shown), thus confirming the results obtained through semi-quantitative RT-PCR. Hence, Emnhr1 is expressed in all larval stages that are involved in the infection of the intermediate host.
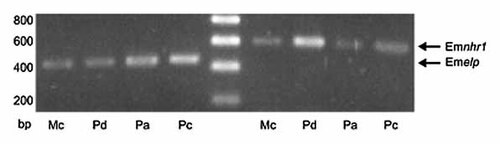
Expression of Emnhr1 in E. multilocularis larval stages. Total RNA was isolated from in vitro-cultivated metacestode vesicles (Mc), dormant (Pd) and low-pH/pepsin-activated protoscoleces (Pa) and in vitro cultivated primary cells (Pc). Total RNA was reverse transcribed and the resulting cDNA was then used as a template for PCR using primers specific for Emnhr1 and the constitutively expressed control gene Emelp [Brehm et al., 2003]. PCR products were run on a 1% agarose gel and stained with ethidium bromide. DNA size marker bands are indicated to the left.
We also analysed the transcriptome dataset for expression profiles of all other putative Echinococcus NRs listed above and found constitutive expression for the majority, at least in the four stages analysed (see Supplementary Table I). A clear differential expression pattern was observed for NRs encoded on contigs 2822 (only expressed in metacestode), 2878 (absent in metacestode vesicles) and 3452 (only expressed in protoscoleces). For two putative NRs (encoded on 1680 and 1798), no or only very weak expression was detected in the larval stages (supplementary Table I).
Protein–Protein Interactions of EmNHR1
NRs typically undergo direct interactions with other cellular proteins such as NR co-activators and co-repressors as well as transcription factors of other families [Aranda and Pascual, 2001]. To identify possible interaction partners of EmNHR1, we employed the yeast two-hybrid system. We had previously reported the identification and characterisation of EmSkip, a member of the SNW/SKIP family of transcriptional co-regulators [Gelmedin et al., 2005] and first used translational EmSkip fusions with the Gal4-LBD and the Gal4-DBD in co-transformation experiments with Gal4-fusions of full-length EmNHR1 (EmNHR1-FL) as well as Gal4-fusions of the EmNHR1 DBD and LBD. However, as shown in Figure 6, no combination resulted in positive interaction, indicating that EmSkip is not involved in Echinococcus signalling pathways together with EmNHR1.
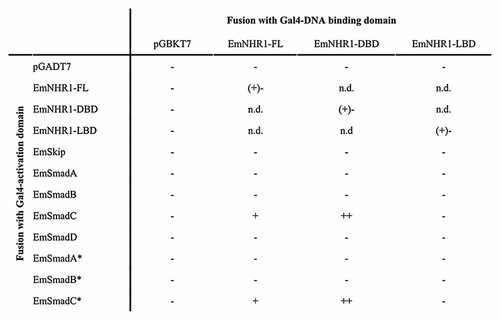
Analysis of EmNHR1 interactions with EmSkip and EmSmads. Translational fusions were generated for the Gal4-activation domain (AD; vector pGBKT7) with the full-length EmNHR1 (EmNHR1-FL), the EmNHR1 LBD and the EmNHR1-DBD. Double transformants of these constructs (yeast strain AH109) together with empty vectors (pGADT7) or with different fusions of the Gal4 DNA-binding domain (DBD; vector pGADT7) with EmSkip [Gelmedin et al., 2005], EmSmadA, EmSmadB [Zavala-Gongora et al., 2003], EmSmadC and EmSmadD [Zavala-Gongora et al., 2008] were generated. An asterisk marks the activated forms of EmSmadA, EmSmadB and EmSmadC [Zavala-Gongora et al., 2003, 2008]. Positive control (pGADT7-T × pGBK-53) and negative control (pGADT7-T × pGBK-Lam) were performed as described in the MATCHMAKER manual (Clontech). Growth of colonies was assessed according to the MATCHMAKER manual (Clontech). −, + and ++ indicate no growth or growth under medium and high stringency conditions. (+)− indicates slight growth under medium stringency conditions, which is blocked by 7.5 mM 3-AT treatment. Yeast two-hybrid analyses in which full-length or parts of EmNHR1 were fused with the Gal4 AD and tested against the other Echinococcus proteins as Gal4 DBD fusions yielded identical results (data not shown).
Another group of NR interaction partners are Smad transcription factors, which are downstream signalling components of TGF-β/BMP signalling in all metazoans [Pendaries et al., 2003]. We had previously characterised four different Smad factors in E. multilocularis of which two represented receptor regulated Smads (R-Smads) of the TGF-β/activin pathway (AR-Smads; EmSmadA, EmSmadC), one a R-Smad of the BMP-pathway (BR-Smad; EmSmadB) and one a co-regulatory Smad (Co-Smad; EmSmadD) [Zavala-Gongora et al., 2003, 2008]. In the yeast two-hybrid system, we therefore tested Gal4-fusions of each of those Smads against EmNHR1-FL as well as the EmNHR1 LBD and DBD. R-Smads are phosphorylated, and thereby activated, by upstream TGF-β type I receptors at a conserved C-terminal pC-X-pC motif which can result in differential interactions with other proteins [Zavala-Gongora et al., 2003, 2008]. For the three Echinococcus R-Smads, we therefore also tested activated forms that had been produced by replacement of the C-terminal Cys residues through phospho-mimetic Glu [Zavala-Gongora et al., 2003, 2008]. As shown in Figure 6, a clear interaction could be observed between EmSmadC (both activated and non-activated) and EmNHR1-FL which was even more pronounced if only the EmNHR1 DBD was used. This indicated a possible cross-talk between NR signalling, through EmNHR1, and TGF-β/activin signalling, through EmSmadC, in E. multilocularis.
The Effect of Serum on EmNHR1-LBD Dimerisation
We were finally interested in the identification of possible ligands for EmNHR1 and whether these might be derived from the host. All members of the HR-96/DAF-12 family identified so far have cholesterol-derived compounds as their ligands [Mooijaart et al., 2005] and for the detection of respective ligand–receptor interactions, a modified version of the yeast two-hybrid system had already frequently been used [Kakizawa et al., 1997; Lezzi et al., 2002; Oftedal et al., 2005]. In this modified yeast two-hybrid, the LBD of NRs is fused to the Gal4-DBD or to the Gal4-AD and double transformants are incubated in the presence of potential ligands. In the presence of non-cognate compounds, no interaction is detected, whereas in the presence of cognate ligand, the NR LBDs form dimers that result in positive readout [Kakizawa et al., 1997; Lezzi et al., 2002; Oftedal et al., 2005]. We therefore produced translational fusions of the EmNHR1 LBD with the Gal4-DBD or with the Gal4-LBD and tested a number of possible ligands such as testosterone, 17β-estradiol, progesterone, cortisone and ecdysone for stimulating activity. However, although all these compounds resulted in overall elevated β-galactosidase activities in yeast double transformants, none produced a statistically significant effect.
We then tested whether host serum could induce dimerisation of the EmNHR1 LBD and, as shown in Figure 7, observed significantly elevated β-galactosidase activities in the presence of 5% human or bovine serum, which was even more pronounced in the presence of higher serum concentrations (data not shown). Likewise, 20% hydatid fluid from in vitro-cultivated metacestode vesicles had a significant effect. Taken together, these results indicated that host serum contains at least one ligand that can be recognised by EmNHR1. When a truncated version of the EmNHR1 LBD was used (lacking parts of the C-terminal LBD region), no elevated β-galactosidase was measured (Fig. 7), indicating that the complete LBD was necessary for ligand-dependent dimerisation of EmNHR1.
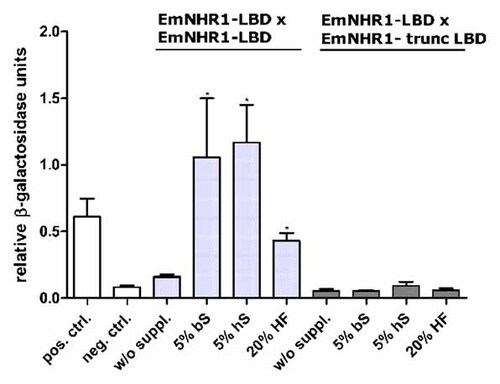
Comparative β-galactosidase liquid assays to determine EmNHR1-LBD dimerisation. Yeast cells (strain Y187) were transformed with fusion constructs of the Gal4 AD and Gal4 DBD with the full-length EmNHR1 LBD or with the EmNHR1 LBD carrying a deletion of the C-terminal LBD part (EmNHR1-trunc LBD). The cells were then cultured over night in selective SD-Leu/-Trp medium either without supplement (w/o suppl.) or in the presence of 5% bovine serum (5% bS), 5% human serum (5% hS) or 20% E. multilocularis hydatid fluid (20% HF). pGAD-ElpN/pGBK-ElpC (positive control [Hubert et al., 2004]; pos. ctrl.) and pGADT7-T/pGBK-Lam (negative control; neg. ctrl) served as controls. The statistical evaluation of three independent measurements of β-galactosidase activity (n = 3) is shown (error bars are indicated). Student's t-test (two-tailed): *P < 0.005.
DISCUSSION
Parasitic platyhelminths express a number of evolutionarily conserved signalling systems by which they are able to sense host-derived peptide hormones and cytokines such as insulin, EGF or TGF-β [Dissous et al., 2006; LoVerde et al., 2007; Brehm, 2010b]. In principle, these parasites should also be able to respond to lipophilic host hormones and in the case of cestodes of the genus Taenia, evidence for an influence of steroid hormones on in vitro parasite development was already obtained [Escobedo et al., 2004, 2010]. Corresponding platyhelminth receptor molecules of the NR family have so far exclusively been described in schistosomes [Wu and LoVerde, 2010] although all of these still have to be classified as ‘orphan receptors’ since no specific ligands have yet been identified. In this work, we extend the analyses of platyhelminth NRs for the first time to cestodes. The receptor we have characterised, EmNHR1, displays all structural features of members of the NR family and contains all highly conserved residues at the corresponding positions. Structurally, EmNHR1 clearly belongs to the DAF-12/HR-96 subfamily of NRs which is considered to be an evolutionarily conserved receptor group that regulates cholesterol homeostasis and longevity in diverse metazoans [Mooijaart et al., 2005; Horner et al., 2009; Bujold et al., 2010]. In C. elegans, DAF-12 binds Δ4 and Δ7 dafachronic acids as ligands which are bile-acid-like compounds that derive from cholesterol, and crucially affects the expression of a set of genes that is necessary for dauer entry under unfavourable growth conditions [Gerisch et al., 2007]. Likewise, Drosophila HR-96 is thought to be activated by cholesterol derivatives and acts as a sentinel for low cholesterol concentrations that induces the expression of genes necessary for uptake and processing of dietary cholesterol [Horner et al., 2009; Bujold et al., 2010]. In mammals, the NRs with highest homologies to DAF-12/HR-96 are the liver X receptors (LXRα, LXRβ), the ligands of which are oxysterols, which are oxidised cholesterol derivatives that serve as intermediary substrates in steroid hormone and bile-acid synthesis [Peet et al., 1998; Mooijaart et al., 2005]. Based on the homologies of the NHR1 LBD to the LBDs of all those NRs, we suggest that the EmNHR1 ligand is also a cholesterol derivative. However, whether EmNHR1 indeed fulfils a similar role in E. multilocularis as DAF-12 or HR-96 in C. elegans and Drosophila, respectively, still requires further experimentation since the parasite genome also encodes two additional NRs with LBD homologies to DAF-12/HR-96 and LXRs. One of these, a member of the invertebrate-specific 2DBD-NR family, is present on contig 2681 and the second (contig 3309) does not only show LBD homologies to HR-96 but is also, like EmNHR1, a member of the CESCKA subfamily (Table I). In contrast to EmNHR1, these two NRs have very close homologs in S. mansoni, Sm2DBDβ and SmHR96α (also called SmCAR for ‘constitutive androgen receptor’ [Hu et al., 2006a]), which would be expected for an evolutionarily conserved signalling system. As an alternative to an Echinococcus-internal ligand for EmNHR1 that arises from cholesterol metabolism, the actual ligand for this NR could also be provided by the host, which is supported by our experiments demonstrating that human and bovine serum contain a stimulus for EmNHR1-LBD dimerisation. The fact that hydatid fluid also stimulated EmNHR1 dimerisation does not argue against the hypothesis of a host-derived ligand for EmNHR1 since this compartment contains a large number of host serum components that are transported into the hydatid fluid by as yet uncharacterised mechanisms [Bernthaler et al., 2009; Monteiro et al., 2010]. Serum is indeed an important component in all cultivation media that support E. multilocularis development in vitro [Spiliotis et al., 2004, 2008, 2010; Spiliotis and Brehm, 2009] and, at least in our hands, all attempts to cultivate the parasite under serum-free conditions or in media deprived of serum lipid components failed so far. EmNHR1 could thus be a sensor for the presence of the parasite in this compartment, possibly up-regulating a set of genes that are required to take up cholesterol or cholesterol-like compounds from the medium. Unfortunately, the yeast two-hybrid system used in our study is particularly designed to detect interactions between NR LBDs and steroid hormones but has never been used to detect similar interactions for oxysterols, which undergo complex modifications within serum and are usually bound to lipoprotein. In future investigations, it might thus be necessary to either use highly sensitive, luciferase-based cellular assays or direct in vitro binding assays on the purified LBD to identify the cognate ligand of EmNHR1.
As concerning downstream target genes for EmNHR1 we demonstrated that this NR, together with the HR-96-like NR encoded on contig 3309, contains a sequence context around the P-box that is highly similar to the DNA recognition α helix of DAF-12 and HR-96, suggesting that they all recognise similar HREs. DAF-12 target genes of C. elegans typically contain hexameric DR5 direct repeats that are separated by 5 nt (consensus: AGTTCA-n5-AGTGCA [Shostak et al., 2004]) and a similar sequence serves as HRE for Drosophila HR-96 (AGTGCA-n5-GTGTCA [Fisk and Thummel, 1995]). Interestingly, in a preliminary bioinformatic approach we could identify similar sequences in the promoter region of an Echinococcus ortholog to the Niemann-Pick disease Type C1 gene (NPC1; present on contig 3045) that encodes a member of the patched protein family (E-value of 1E−177 when compared to Drosophila NPC1). In Drosophila, this protein is required for sterol absorption in the midgut epithelium under control of HR-96 [Voght et al., 2007], while the respective ortholog in mammals is regulated by the DAF-12/HR-96 ortholog LXR [Peet et al., 1998]. This indicates that, like in Drosophila and mammals, NPC1 could also be regulated by the DAF-12/HR-96 orthologs in Echinococcus. Further possible targets for the CESCKA family NRs in Echinococcus that carry DR5-like repeats in the promoter are genes encoding a Fyn-like kinase and a pou-domain-like transcription factor (data not shown), and we are currently testing whether EmNHR1, in comparison to the Echinococcus SmCAR-like NR, can bind to the respective promoter sequences. Since, in S. mansoni, SmCAR binds to its cognate sequences as a hetero-dimer with SmRXR1 [Hu et al., 2006b], these studies will also address the possibility that EmNHR1 (or the Echinococcus SmCAR ortholog) might form hetero-dimers with the NR encoded on contig 3440, which is no close ortholog of the schistosome RXRs but at least displays homologies to SmRXR2 in the LBD (Table I).
Another finding of this study that is important for EmNHR1 downstream signalling mechanisms is the observed interaction between the EmNHR1 DBD and the AR-Smad EmSmadC, which forms part of the parasite's TGF-β/activin signalling pathway [Zavala-Gongora et al., 2008]. In C. elegans, it is well established that TGF-β and insulin signalling pathways converge on DAF-12 by inducing the production of dafachronic acid through DAF-9 (cytochrome p450 [Gerisch et al., 2007]). To our knowledge, direct interactions between DAF-12 or HR-96 and components of the TGF-β signalling pathways have not yet been demonstrated in C. elegans or Drosophila. In mammals, on the other hand, at least the distantly related vitamin D receptor (VDR) has been shown to interact with Smad factors and the LXR can form complexes with both AR-Smads (Smad2/3) using the NR co-activator RAP250 as a bridging molecule [Antonson et al., 2008]. Hence, cross-communication between NRs that regulate cholesterol homeostasis in vertebrates and invertebrates with the respective TGF-β signal transducers appear to be another conserved feature of these signalling systems, although the precise molecular mechanisms might differ in so far as TGF-β signalling either induces the production of cognate NR ligands or that downstream Smad transcription factors, either directly or indirectly, interact with the NR component.
Our bioinformatic analyses on the Echinococcus genome revealed that this cestode expresses a set of NRs that broadly overlaps with that of S. mansoni and S. japonicum. Of the 17 NRs that are encoded by the Echinococcus genome, 11 displays clear homologies within the DBD and the LBD to orthologous factors in schistosomes, indicating similar function. For one additional potential Echinococcus NR, encoded on contig 2891, we did not obtain prediction of an LBD but neither is an LBD predicted for the longest available EST sequence of the closest S. mansoni ortholog, SmHR96β, again indicating similar structure and function. Notable differences between both species are that no clear SmRXR ortholog is predicted in the E. multilocularis genome which, instead, carries a gene with certain homologies to SmRXR2 in the LBD but a different DBD. Whether this NR is able to undergo hetero-dimer formation with other Echinococcus NRs, as typical for this type of receptors, remains to be established. Furthermore, two branches in which schistosomes express three relatively closely related receptors (SmTLL–SmDSF–SmPNR; SmTRα–SmTRβ–SmRAR-like) are only represented by one ortholog in Echinococcus. The cestode, on the other hand, expresses at least two receptors that do not have orthologs in schistosomes. One of these in encoded on contig 2878 (LBD homologies to Drosophila HR-78), and the second is EmNHR1.
Mining of the available transcriptome dataset revealed that the majority of the predicted NRs is either constitutively expressed in all four larval stages analysed so far or shows preferential expression in the protoscolex (contig 3425) or the metacestode (contig 2822). For two predicted NRs (contigs 1680, 1798), no significant expression was detected in any larval stage. We cannot rule out the possibility that these are pseudogenes, although we did not detect an accumulation of nonsense mutations in the reading frames. An alternative explanation would be that these genes are preferentially expressed in the adult worm (e.g., during sexual differentiation), since for this stage no transcriptome data are presently available.
It should be noted that Escobedo et al. [2004] already claimed the identification of ‘estrogen receptors’ (α and β isoforms) and an ‘androgen receptor’, which are all members of the NR family, in the cestode T. crassiceps. Recently, this group also reported on the presence of a ‘progesterone receptor’ in T. solium that apparently displayed high homologies to vertebrate members of the progesterone receptor subfamily of NRs [Escobedo et al., 2010]. None of these studies provided the actual sequences of the cestode NRs. Instead, primers specifically designed against the DBDs of murine estrogen, androgen and progesterone receptors were employed in PCR amplification from Taenia parasite material, and a positive reaction was taken as evidence that the respective receptors also exist in these cestodes [Escobedo et al., 2004, 2010]. Our analyses of the E. multilocularis genome did not reveal the presence of such receptors, nor are NRs with high homology to vertebrate steroid receptors present in the genomes of schistosomes [Wu and LoVerde, 2010] or planarians (our own unpublished analyses). There are two possible explanations for these discrepancies. On the one hand, cestodes of the genus Taenia could significantly differ from all other flatworms in their set of NR encoding genes, being equipped with additional members that display considerable homologies to vertebrate sex steroid receptors (possibly acquired by horizontal gene transfer). On the other hand, the amplified PCR products could have resulted from host contamination that was present in the Taenia parasite material, which was directly taken out of mammalian hosts [Escobedo et al., 2004, 2010]. We consider the second explanation likelier, which does, of course, not exclude that mammalian sex steroids might interact with one of the ‘flatworm-typical’ NRs that we have identified in E. multilocularis. If so, the NR encoded on contig 3442 (ortholog of SmFTZ-F1) is a promising candidate since in the LBD, this receptor displays the highest homologies of all identified cestode NRs to the LBDs of vertebrate estrogen and androgen receptors (∼26% identical and ∼45% similar amino acids).
Taken together, in this study we have made a first step towards the characterisation of nuclear hormone receptor signalling in cestodes. We demonstrate that the model cestode E. multilocularis expresses a set of NRs that broadly overlaps with that of the related flatworms S. mansoni and S. japonicum, but also contains several NR encoding genes that are unique to this parasite (and probably also to other cestodes). For one of these NRs, EmNHR1, we furthermore demonstrated cross-communication with TGF-β signalling components and that mammalian serum, a necessary component for larval cestode development, contains a cognate ligand. EmNHR1 could thus have a role in hormonal host–parasite cross-communication during an infection. On the basis of our results, further investigations into the role of NR signalling in cestode development and host–parasite interaction will be facilitated.
Acknowledgements
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFB; grant SFB 479) and the Wellhöfer Foundation (to K.B.). S.F. was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg. The authors wish to thank Monika Bergmann and Dirk Radloff for excellent technical assistance. Sequence data of the E. multilocularis and E. granulosus genome sequencing projects have been produced by the Parasite Sequencing Group at the Sanger Institute (Matthew Berriman) and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/Echinococcus.




