Chloride intracellular channel 5 modulates adipocyte accumulation in skeletal muscle by inhibiting preadipocyte differentiation
Abstract
Intramuscular fat, the total lipid deposited within skeletal muscle, has been regarded as a potential factor responsible for meat quality in animal production and insulin resistance in humans. The objective of present study was to identify candidate genes which control intramuscular fat accumulation through using animal models. PIC pigs (lean-type) and Rongchang pigs (obese-type) were used. By scanning the mRNA samples of longissimus dorsi muscle with Affymetrix Gene-Chip microarray technology, sus scrofa chloride intracellular channel 5 (CLIC5) was isolated, and its mRNA abundance and protein expression level were reversely related with the intramuscular fat content of pigs. Furthermore, over-expression of CLIC5 dramatically increased the proliferation of 3T3-L1 preadipocytes, while inhibited adipocytic differentiation accompanied by the down-regulation of c/EBPα, LPL, and PPARγ protein. Our results suggest that CLIC5 might be a crucial regulator of adipose accumulation in skeletal muscle of pigs. J. Cell. Biochem. 110: 1013–1021, 2010. © 2010 Wiley-Liss, Inc.
Abbreviations used:
CLIC, chloride intracellular channel; FBS, fetal bovine serum; DMEM, Dulbecco's modified Eagle's medium; PPARγ, peroxisome proliferator-activated receptor γ; ADD1, adipocyte determination and differentiation dependent factor-1; c/EBPα, CCAAT enhancer binding protein α; LPL, lipoprotein lipase; ESTs, expressed sequence tags.
Skeletal muscle is a complex tissue demonstrating considerable plasticity in its capacity to modify the contribution of energy derived from the macronutrients. Intramuscular fat, the total lipid accumulation within skeletal muscle, is located mainly in perimysium and to a lesser extent in muscle cells [Wood, 1990]. It refers to triacyglycerol occurring primarily within adipocytes between bundles of muscle fibers [Harper and Pethick, 2004]. Myocytes do accumulate some fat as droplets, but this intramyocellular lipid is not a major contributor to the total intramuscular fat pool or marbling [Dagenais et al., 1976; Essen-Gustavsson et al., 1994; Gondret et al., 1998]. Intramuscular fat is closely related to sensory properties of meats in animal production [Wood, 1990; Huff-Lonergan et al., 2002] and insulin resistance in humans [Virkamaki et al., 2001; Goodpaster and Wolf, 2004].
Muscle lipid accumulation in humans has negative impacts on health; on the other hand, it is the main factor positively affecting meat edible quality including meat tenderness, juiciness, and taste [Cameron et al., 2000]. However, for many years, a major objective of swine industry has been to increase the lean to fat ratio of pork [Kouba and Bonneau, 2009]. As a result, modern pig genetic-types deposit less fat and are much leaner at market weight than traditional breeds [Solanes et al., 2008]. Since the phenotypes of lean and intramuscular fat content are negatively correlated [Hovenier et al., 1992], the intramuscular fat content of the pig meat is below the level considered to be optimum in terms of the sensory properties of pork [Gerbens et al., 1999; Morlein et al., 2005]. Intramuscular fat and subcutaneous fat are influenced by different genes [Janss et al., 1997] suggesting that intramuscular fat level could be modulated through regulating key genes without increasing other fat depots. Thus, it is necessary to identify candidate genes that might contribute to alter intramuscular fat content in skeletal muscle. However, such genes have not yet been isolated [Jiang et al., 2007]. Chloride intracellular channels (CLICs) are relatively small proteins, and the roles of which as ion channels are poorly understood. To date, CLIC family was investigated through reconstitution in planar bilayer in vitro to form channels [Tulk et al., 2002; Warton et al., 2002; Littler et al., 2005; Singh et al., 2007], and very little is known about the physiological roles of CLICs in vivo. Thus, they may have additional molecular functions, and it is yet unclear that whether they associate with adipose accumulation.
Breed is considered as an important factor influencing the intramuscular fat content in swine [Cagnazzo et al., 2006]. Rongchang pigs, an indigenous breed that distributes mainly in the southwest area of China, have almost three times the intramuscular fat levels of the European hybrid pigs [Zheng, 1986; Lu et al., 2008]. Thus, the objective of present study was to identify genes that were differently expressed in the longissimus dorsi muscle between Chinese indigenous breed (Rongchang obese-type pigs) and European hybrid pigs (PIC lean-type pigs) in order to identify candidate genes regulating intramuscular fat accumulation.
MATERIALS AND METHODS
Animals
All procedures used in this experiment were approved by the Institute of Animal Care and Use Committee of China Agricultural University. There were six pens housing each breed and four barrows in each pen. During the growth phase of the experiment, all pigs had ad libitum access to a standard corn–soybean diet and water. Pigs from the two breeds received the identical treatment throughout the study. One barrow was randomly selected for slaughter from each pen at an average live weight of 60 kg (n = 6) and 90 kg (n = 6). The barrows were slaughtered using electric stunning followed by exsanguination.
Sample Collection
Samples of the longissimus dorsi muscle and subcutaneous adipose tissue were taken from the left side of the carcass at the last rib immediately after exsanguination. For longissimus dorsi muscle samples, all visible intermuscular (between muscles) fat was removed. All tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until analyses.
Backfat Depth and Intramuscular Fat Content
Backfat depth was measured at the 6th and 7th thoracic rib of the carcass using Vernier Calipers (Mitutoyo Company, Kawasaki, Japan). The total lipid content in longissimus dorsi muscle was determined following the method of the Association of Official Analytical Chemists [AOAC, 1990].
Interestingly, two carcasses of Rongchang pigs (one at 60 kg and another at 90 kg) had an intramuscular fat content over two times higher than the rest Rongchang pigs even though their backfat depth was similar. It is a rare phenomenon. Hence, these two special Rongchang pigs were analyzed separately, which provided us the pigs with a hierarchy of intramuscular fat content ranging from PIC-lean type pigs, Rongchang pigs, and Rongchang pigs with high intramuscular fat.
Total RNA Extraction
Total RNA was isolated from the longissimus dorsi muscle and subcutaneous adipose tissue using RNeasy Fibrous Tissue Mini Kit and RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany), respectively. The integrity of the RNA was verified by electrophoresis in a 1% agarose gel stained with ethidium bromide. The quality and quantity of RNA were determined by ultraviolet spectroscopy using a NanoDrop® ND-1000 (Thermo Fisher Scientific, Wilmington, DE).
Microarray
Total RNA of muscle tissue from three slaughtered pigs of each breed and the special Rongchang pig at each stage was individually hybridized with the GeneChip Porcine Genome Array (Affymetrix, Santa Clara, CA), which contains 23,937 probes (23,256 transcripts) representing 20,201 sus scrofa genes. Total RNA (10 µg) was reverse transcripted to generate first-strand cDNAs using the SuperScript III (Invitrogen, Carlsbad, CA) with T7-(d7) 24 primer. Double-stranded cDNA was purified according to the manufacturer's instructions (Affymetrix). Biotinylated cRNA was synthesized using the Enzo Bio Array High Yield RNA Transcript Labeling Kit (Enzo Life Sciences, Inc., Plymouth Meeting, PA). Labeled cRNA was fragmented and hybridized to the GeneChips microarrays using the Affymetrix Automated GeneChip System. Twelve micrograms of biotinylated target cRNA were hybridized per microarray. After hybridization, washing, and staining, the samples were scanned using the GeneChip-Scanner 3000 (Affymetrix). Quantitative analysis of microarray hybridization was performed using Affymetrix Micro Array Suite 5.0-Specific Terms (Statistical Algorithms) GCOS (Affymetrix GeneChip Operating Software) Version 1.4. A differentially expressed gene was defined as a variation in gene expression no less than twofolds.
Molecular Cloning and Sequencing
Homology-based polymerase chain reactions were employed under the action of Pfu DNA polymerase. A poly-dA tail was added to the 3′-end of the primary sequence using a terminal kit (Takara Bio, Inc., Dalian, China). cDNA PCR products of interest were purified with the QIAquick PCR Purification Kit (Qiagen). Sequencing was conducted using an Applied Biosystems 3730 DNA Sequencer (AuGCT Biotechnology, Beijing, China). Then, cDNA sequences were analyzed using the software programs EDITSEQ and MEGALIGN, respectively (DNASTAR, Madison, WI).
Semi-Quantitative RT-PCR
Total RNA of muscle and adipose tissue from all the slaughtered pigs of each breed was used. Primers for porcine CLIC5 (sense: 5′-TTG CTG GAT GTG AGT CTG CT-3′, antisense: 5′-GCC TAT CAA ATG TCC TCC TC-3′) and β-actin (sense: 5′-TGC GGG ACA TCA AGG AGA AG-3′, antisense: 5′-AGT TGA AGG TGG TCT CGT GG-3′) were synthesized by the Shanghai Sangon Biological Engineering Technology Company (Shanghai, China). Amplification conditions were 95°C 1 min, 28 cycles of 30 s at 95°C, 30 s at 62°C, 1 min at 72°C followed by 8 min at 72°C and carried out in a Peltier Thermal Cycler PCR System (PTC-200, MJ Research, Waltham, MA). PCR products were fractionated by electrophoresis and verified by sequencing. β-Actin was used as inner control. Alpha Imager 2200 (Alpha Innotech Corporation, San Leandro, CA) software was used to determine the density of the cDNA bands.
Cell Culture
3T3-L1 preadipocytes were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) fetal bovine serum (FBS) (both from Gibco, Grand Island, NY), in a 5% CO2 incubator at 37°C.
Vector Construction and Transfection of 3T3-L1 Preadipocytes
To construct the vector of pcDNA3.1-CLIC5, the coding sequence of CLIC5 was subcloned into the EcoRI and the HindIII sites of the pcDNA3.1-Myc/His (B) expression vector (Invitrogen) using oligonucleotides 5′-CGG AAT TCA TGA CGG ACG CGG CGA CGG C-3′ and 5′-CCC AAG CTT TCA GGA GCG GCT CAG GCG CT-3′. Then the cells were transfected using Lipofectamine2000 (Invitrogen). Before transfection, the cells were grown to 90% confluence in six-well plates, and 2 µg of DNA and 6 µl of liposome reagent were premixed in FBS-free DMEM medium followed by 20 min of incubation at room temperature. Then, cells in each well were incubated with 500 µl of the DNA/liposome mixture. Six hours later, the mixture was substituted by fresh DMEM medium with 10% FBS. Forty-eight hours after transient transfection, neomycin G418 (Amaresco, Solon, OH) was added to the medium (500 µg/ml) to select transfected cells. Western blotting analysis was used to confirm the expression of CLIC5 in transfected cells. The colonies expressing the significantly higher level of CLIC5 protein than that of the control were selected for further study.
Proliferation Analysis of the Transfected 3T3-L1 Preadipocytes
The cells transfected with or without the CLIC5 gene were seeded in 96-well culture plates at a density of 5 × 102 cells/well and supplemented with 10% FBS with 200 µg/ml G418. Ten microliters of the CCK-8 solution (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H-tetrazolium, monosodium salt; Dojindo Laboratories, Kumamoto, Japan) was added to each well, which also contained 100 µl DMEM. After 1 h incubation at 37°C, the absorbance at 450 nm of the suspension was measured.
Cell Cycle Analysis of the Transfected 3T3-L1 Preadipocytes
Flow cytometry was used to examine the cell cycle. Before analysis, cells were serum deprived for 48 h to synchronize the cell cycle. Then cells were incubated with DMEM supplemented with 10% FBS for various times (0, 12, 18, and 24 h). Cells were harvested and re-suspended in a phosphate-buffered saline (PBS)/ethanol mixture (30/70%), treated with 1 mg/ml RNAase A, and then mixed with 50 µg/ml propidium iodide (Sigma). DNA content was then analyzed with a FACS Calibur System (Becton Dickinson, Boston, MA).
Differentiation of 3T3-L1 Preadipocytes
Two days after complete confluence (day 0), cells were cultured in DMEM containing 10% FBS, 0.5 mM 1-methyl-3-isobutylxanthine, 1 µM dexamethasone, and 5 µg/ml insulin (all from Sigma) for 48 h. From days 2 to 4, the complete medium was supplemented with 100 nM insulin only, followed by culturing in DMEM containing 10% FBS up to day 8. Then the cells were washed with PBS three times and fixed for 5 min with 4% paraformaldehyde. Fixed cells were incubated with Oil red O for 2 h and then hematoxylin for 2 min at room temperature. Stained lipid droplets in cell monolayers were visualized by light microscopy and photographed.
Western Blotting Analysis
Tissues or harvested cells were treated with RIPA lysis buffer (JingKeHongDa Biotechnology, Beijing, China) to isolate the protein. About 30 µg of the protein extract was size-fractionated by gel electrophoresis and transferred to nitrocellulose membranes. Blots were incubated with rabbit CLIC5 polyclonal antibodies (AVIVA Systems Biology, San Diego, CA) at a 1:4,000 dilution, or adipocyte determination and differentiation dependent factor-1 (ADD1) monoclonal antibodies, goat anti-CCAAT enhancer binding protein α (c/EBPα), rabbit anti-lipoprotein lipase (LPL) polyclonal antibody, and mouse anti-peroxisome proliferator-activated receptor γ (PPARγ) (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:200 dilution. Then, the membranes were rinsed in TBST (20 mmol/L Tris, 500 mmol/L NaCl, 0.1% Tween-20, pH 7.6), and incubated with peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (Santa Cruz Biotechnology) for 1 h at a dilution of 1:5,000. The bands of the protein were visualized using a chemiluminescent reagent (Pierce, Rockford, IL). β-Actin antibody (Santa Cruz Biotechnology) diluted at 1:1,000 was used to normalize the amount of proteins. Alpha Imager 2200 (Alpha Innotech Corporation) software was used to determine the density of the protein bands.
Statistical Analysis
The Cluster method [Shen et al., 2005; Zou et al., 2009] was carried out to assign the chip data of the Rongchang pigs with high intramuscular fat into the proper group. The statistical analysis was done by unpaired t-test or one-way ANOVA. The level of significance was set at P < 0.05.
RESULTS
As expected, Rongchang pigs had significantly higher intramuscular fat and backfat depth than the PIC lean-type pigs (P < 0.01, Table I). During the experiment, two Rongchang pigs whose intramuscular fat content was over two times than the rest Rongchang pigs were separated. However, the backfat depth of these two Rongchang pigs was nearly equal to the other Rongchang pigs. Therefore, these two Rongchang pigs were considered as a separated group for data analyses.
| Slaughter weight (kg) | Items | PIC lean-type pigs | Rongchang obese-type pigs | P-value | Rongchang pigsa with high intramuscular fat |
|---|---|---|---|---|---|
| 60 | Backfat depthb (mm) | 18.00 ± 0.71 | 31.40 ± 1.87 | <0.0001 | 37.10 |
| Intramuscular fat (%) | 1.26 ± 0.16 | 3.38 ± 0.46 | <0.001 | 8.13 | |
| 90 | Backfat depth (mm) | 26.90 ±1.17 | 44.40 ± 1.93 | <0.0001 | 46.00 |
| Intramuscular fat (%) | 1.15 ± 0.15 | 4.64 ± 0.39 | <0.0001 | 9.05 |
- a The Rongchang pigs whose intramuscular fat was over two times higher than the rest Rongchang pigs even though their backfat depth was similar. Data for these pigs were not considered in the statistical analysis because of their limited numbers.
- b Backfat depth was measured at the 6th and 7th thoracic rib of the carcass using Vernier Calipers.
Identification of Differentially Expressed Genes
Through microarray assay, we acquired several expressed sequence tags (ESTs) whose expression exhibited similar or opposite patterns corresponding to the intramuscular fat content of the pigs. Notably, the microarray-based mRNA expression of EST BP439390 and BX666230 (GenBank Accession No.) in PIC pigs was significantly higher (P < 0.05) than that of Rongchang pigs at 60 and 90 kg. For both ESTs, mRNA abundance was inversely related to intramuscular fat content (Table II).
| Slaughter weight (kg) | EST | PIC lean-type pigs | Rongchang obese type pigs | t-test | Ratioa (lean vs. obese) | Rongchang pigsbc with high intramuscular fat |
|---|---|---|---|---|---|---|
| 60 | BP439390 | 966 | 222 | 0.01 | 4.35 | 120 |
| BX666230 | 5,198 | 2,290 | 0.01 | 2.27 | 1,071 | |
| 90 | BP439390 | 1,048 | 127 | 0.03 | 8.21 | 26 |
| BX666230 | 8,576 | 3,968 | 0.02 | 2.16 | 2,725 |
- a A differentially expressed gene was defined as a variation in gene expression no less than twofolds.
- b The Rongchang pigs whose intramuscular fat was over two times higher than the other Rongchang pigs even though their backfat depth was similar.
- c The Cluster method was carried out to assign the chip data of the Rongchang pigs with high intramuscular fat into the proper group and conducted t-test.
After cloning and sequencing, we found that EST BP439390 and BX666230 represented the same gene, and its full-length cDNA shared a high degree of similarity (88%) with human chloride intracellular channel 5A (CLIC5A, GenBank Accession No. NM_016929). The full-length cDNA sequence of the gene was successfully cloned using homology-based and overlapping polymerase chain reactions, and submitted to the GenBank with Accession No. of EU715290 (Fig. 1A). It also shared 97% identity with the DNA sequence of Clone CH242-208015 (GenBank Accession No. CR974564.11) located on porcine chromosome 7. The full-length cDNA of porcine CLIC5 was 3,212 bp, containing 759 nucleotides of open reading frame, predicted encode 252 amino acids with a calculated molecular mass of 32 kDa. Besides, the EST BP439390 and BX666230 were located at the nucleotide 1817 to 2563 and 2720 to 3197, respectively. Sequence analysis using BLASTp revealed that the deduced amino acid sequence of the gene shared a high degree of similarity with human CLIC5A NP_058625 (95.63%) and rat CLIC5 NP_446055 (94.44%), as shown in Figure 1B. Taking these results collectively, we deduced that the novel gene found in the present study is the sus scrofa orthologue of CLIC5.
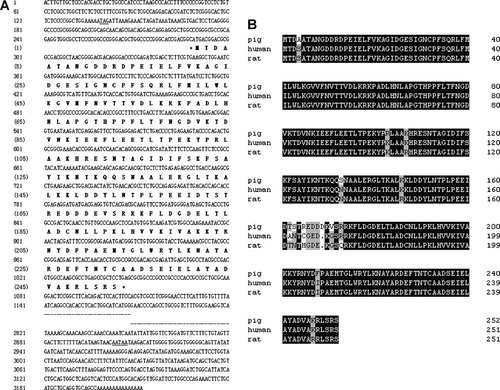
Nucleotide and deduced amino acid sequence of pig CLIC5. A: The amino acid sequence is shown in boldface letters. The underlined nucleotides are the first upstream in frame stop codon and the potential polyadenylation signal (AATAA), respectively. The initiating ATG and the stop codon TGA are marked with a star (*). B: Multiple sequence alignment of the deduced amino acid sequence of porcine CLIC5 compared with human (NP_058625) and rat (NP_446055). Residues highlighted with black shading indicate absolutely conserved amino acids; residues highlighted with gray shading indicate at least the two species sharing the conserved amino acids at that position; residues without shading indicate non-conserved amino acids.
Expression of CLIC5 in Skeletal Muscle and Adipose Tissue
Confirmation of CLIC5 mRNA and protein differential expression in longissimus dorsi muscle from PIC lean type and Rongchang obese type pigs was performed by semi-quantitative RT-PCR and Western blotting. Meanwhile, to determine whether the tendency of CLIC5 expression level in different tissue was the same, subcutaneous adipose tissue was employed. As shown in Figure 2A,B, the mRNA abundance of CLIC5 was significantly elevated in the longissimus dorsi muscle from lean-type PIC pigs compared with obese-type Rongchang pigs at both 60 and 90 kg slaughter weights (P < 0.01). A similar trend was observed in adipose tissue. CLIC5 protein expression level was severely down regulated in the Rongchang pigs comparing with PIC pigs in both skeletal muscle and adipose tissue at different slaughter weights (Fig. 2C,D). Thus, CLIC5 protein levels presented the similar tendency as mRNA abundance.
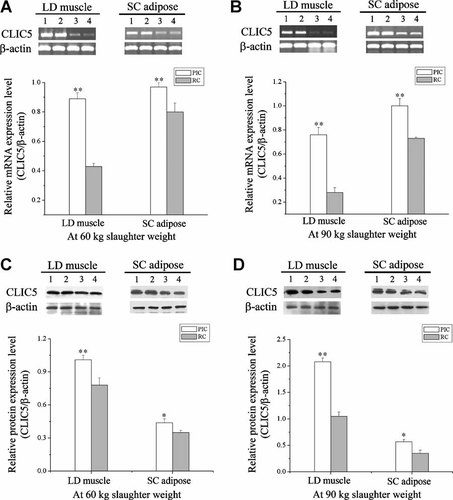
Relative mRNA abundance and protein expression levels of porcine CLIC5 in longissimus dorsi (LD) muscle and subcutaneous (SC) adipose tissue. Total RNA from the samples of 60 kg (A) and 90 kg (B) slaughter weight were reversed transcribed and an aliquot of the cDNA was amplified. Five microliters of PCR product was subjected to electrophoresis in a 1% agarose gel. Western blots containing 30 µg total protein of the samples from the 60 kg (C) and 90 kg (D) slaughter weight were used. β-Actin was used as the internal control. Data represent three independent assays. Lanes 1 and 2: PIC pigs, lanes 3 and 4: Rongchang (RC) pigs. *P < 0.05, **P < 0.01, mean ± SEM (n = 6 for each group).
CLIC5 Promotes Cell Growth
Over-expression of CLIC5 in 3T3-L1 preadipocytes resulted in a remarkably higher proliferation rate compared with controls, while the rate of the cells transfected with an empty vector was almost the same as the controls (Fig. 3A). To elucidate how CLIC5 promoted the proliferation of 3T3-L1 cells, cell cycle of each subclone was studied. More than 80% cells of all cell lines stayed in G0/G1 phase after serum starvation for 48 h. The percentage of CLIC5-expressing 3T3-L1 cells in the S phase increased 18 h after serum stimulation (P < 0.01). Conversely, the percentage of cells transfected with an empty vector or without transfection in S phase at 18 and 24 h after treatment was less than CLIC5 transfected cells (P < 0.01, Fig. 3B).
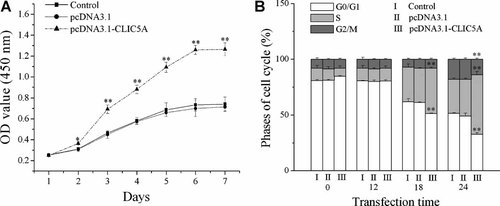
The effect of CLIC5 over-expression on 3T3-L1 cell growth. A: Proliferation of CLIC5 transfected cells in vitro. B: Cell cycle profile of CLIC5 transfected cells. The figure represents one of three independent experiments. *P < 0.05, **P < 0.01, mean ± SEM (n = 6 for each group).
Effects of CLIC5 on Differentiation of 3T3-L1 Preadipocytes
We investigated the effects of CLIC5 on preadipocytes differentiation in 3T3-L1 cells on day 8 after differentiating. Following induced differentiation, numerous fat droplets were observed in the controls and cells transfected with the empty vector. But few lipid droplets appeared in the CLIC5 over-expressing 3T3-L1 cells (Fig. 4). Moreover, over-expression of CLIC5 remarkably down-regulated the protein expression level of c/EBPα, PPARγ, and LPL, but no difference in the expression level of ADD1 was observed (Fig. 5).
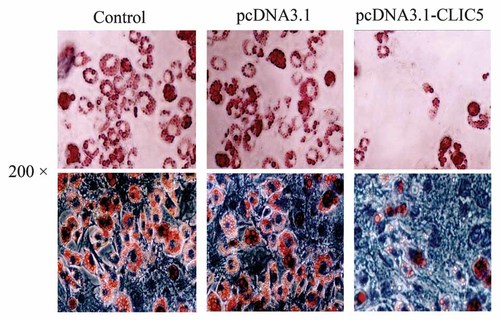
Effects of CLIC5 over-expression on differentiation of 3T3-L1 preadipocytes. Adipocytic differentiation of 3T3-L1 cells transfected with CLIC5, empty vector (pcDNA3.1 Myc/His B) and without transfection were cultured up to day 8. Cells were fixed, stained with Oil red O and hematoxylin (200×) to visualize lipid droplets.
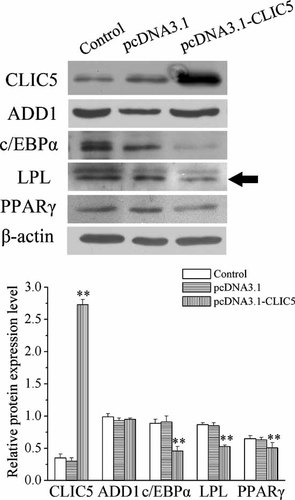
Effects of CLIC5 over-expression on the expression level of adipocyte-specific proteins during preadipocyte differentiation. Cells were collected on day 8 after differentiating. Protein levels were determined by Western blotting. Western blots containing 30 µg total protein of the harvested cell samples were used. The relative expression level of a given protein was caculated after normalization to β-actin expression. Data represent three independent assays. Arrowhead indicated the line used for quantifying. **P < 0.01, mean ± SEM (n = 6 for each group).
DISCUSSION
Pig has always been recognized as a major mammalian model for human biology, due to its similarities to human beings in terms of body size, physiology, and metabolic disease, for example, obesity, diabetes, and heart disease [Lunney, 2007]. The Rongchang pig is an indigenous Chinese breed of pig famous for its high intramuscular fat content and good eating quality [Lu et al., 2008], and the PIC pig is a European hybrid (Large White × Landrace) which has been deliberately selected to increase the lean to fat ratio of its carcass.
Microarray assay was used to identify genes that were differentially expressed in the longissimus dorsi muscle obtained from the three groups of pigs: PIC pigs, Rongchang pigs, and Rongchang pigs with high intramuscular fat content. The sus srofa CLIC5 gene containing EST BP439390 and BX666230 was isolated. CLIC5 belongs to CLIC family including CLIC1-4, CLIC5A, CLIC5B (p64), and CLIC6 (parchorin). Members of the CLIC family share a highly conserved core of ∼230 amino acids. Regarding human CLIC5, alternate splicing results in two different mRNAs that encode proteins of CLIC5A (251 amino acids) and CLIC5B (410 amino acids) with identical C-terminal 238 amino acids [Berryman and Bretscher, 2000; Shanks et al., 2002]. Despite their name, the molecular function and physiological relevance of CLIC proteins is poorly understood. Recently, several articles show that mammalian CLIC5A interacts extensively with cytoskeletal actin [Berryman and Bretscher, 2000; Berryman et al., 2004], display anion channel activities in reconstituted planar lipid bilayer systems [Singh et al., 2007], and are essential for normal inner ear function [Gagnon et al., 2006]. However, it is possible that they have alternative functions unrelated to ion transport. Our results indicated the microarray-based expression level of CLIC5 gene was up-regulated in Rongchang pigs compared to Rongchang pigs with high intramuscular fat content, though their backfat depth were nearly equal. Furthermore, porcine CLIC5 mRNA abundance and protein expression level were both higher in longissimus dorsi muscle or subcutaneous fat of PIC pigs than that of Rongchang pigs, and reversely related with intramuscular fat content. Therefore, we deduced that porcine CLIC5 gene was down-regulated in response to intramuscular fat content.
This is the first report to study the relation between CLIC5 and intramuscular fat. In order to study further, we detected the effects of CLIC5 on preadipocytes proliferation and differentiation. Over-expression of CLIC5 promoted the growth rate of 3T3-L1 cell, and drove more cells into S phase. These data showed that CLIC5 probably facilitated proliferation of 3T3-L1 cells by promoting the replication of genomic DNA. Adipogenesis is regulated by the sequential expression of various transcription factors, such as C/EBPα, PPARγ families, and ADD1 [Rosen and Spiegelman, 2000; Ailhaud and Hauner, 2004]. Preadipocyte differentiation is accompanied by dramatic increases in the fat cell-specific expression of PPARγ and other adipogenic differentiation marker proteins. Those are necessary for the transition of preadipocytes into adipocytes in vitro. Moreover, LPL plays a central role in the control of lipid accumulation [Goldberg, 1996], and is key enzyme involved in the regulation of lipid metabolism. Their expressions were increased during 3T3-L1 preadipocytes differentiation [Gregoire et al., 1998]. The current study showed that over-expression of CLIC5 down-regulated the expression level of c/EBPα, LPL, and PPARγ, but not for ADD1 during the differentiation of 3T3-L1 preadipocytes. ADD1 is involved in a metabolic cascade leading the production of endogenous PPAR ligands [Fajas et al., 1999]. For the present study, CLIC5 did not affect the protein expression of ADD1 during preadipocytes differentiation. Therefore, c/EBPα, LPL, and PPARγ repression more pronounced than that of ADD1.
For the present study, we deduced that elevated expression of CLIC5 could promote the preadipocytes proliferation but suppress differentiation through regulating the key proteins expression level. CLIC proteins are widely expressed in multicellular organisms coexisting in both soluble and integral membrane forms. However, the soluble proteins are poorly understood, and more interest has centered on membrane CLICs. Hence, the interaction between lipid composition of membrane and the biological function of channel proteins may exist. To our knowledge, this study is the first report showing that CLIC5 might play a key role in regulating adipose development. Following work should be conducted to interpret the exact molecular mechanism of CLIC5 in adipogenesis within skeletal muscle.
In summary, we identified porcine CLIC5 gene whose mRNA abundance and protein expression level were reversely related to intramuscular fat content of pigs. CLIC5 may represent a novel regulator that controls preadiopocyte differentiation. A better understanding of its underlying mechanism may provide new insights into the regulation of fat accumulation in skeletal muscle. It would benefit the development of animal production and medical practice.
Acknowledgements
We thank Prof. Zheng Su for microarray data analysis and Prof. Phil Thacker for critical reading of this manuscript.