Over-expression of EDAG in the myeloid cell line 32D: Induction of GATA-1 expression and erythroid/megakaryocytic phenotype
Abstract
Erythroid differentiation-associated gene (EDAG), a hematopoietic tissue-specific transcription regulator, plays a key role in maintaining the homeostasis of hematopoietic lineage commitment. However, the mechanism and genes regulated by EDAG remain unknown. In this study, we showed that overexpression of EDAG in a myeloid cell line 32D led to an erythroid phenotype with increased number of benzidine-positive cells and up-regulation of erythroid specific surface marker TER119. The megakaryocytic specific marker CD61 was also induced significantly. Using a genome-wide microarray analysis and a twofold change cutoff, we identified 332 genes with reduced expression and 288 genes with increased expression. Among up-regulation genes, transcription factor GATA-1 and its target genes including EKLF, NF-E2, Gfi-1b, hemogen, SCL, hemoglobin alpha, beta and megakaryocytic gene GPIX were increased. Silencing of EDAG by RNA interference in K562 cells resulted in down-regulation of these genes. Taken together, EDAG functions as a positive regulator of erythroid/megakaryocytic differentiation in 32D cells associated with the induction of GATA-1 and its target genes. J. Cell. Biochem. 110: 866–874, 2010. © 2010 Wiley-Liss, Inc.
To maintain blood production throughout life, hematopoietic stem cells (HSC) must coordinate alternative fates of self renewal, multilineage differentiation, quiescence, and apoptosis [Kondo et al., 2003; Pina et al., 2008]. A classical view of the hematopoietic hierarchy proposes that multipotent progenitors (MPP) undergo a series of binary decisions resulting in successive dichotomic loss of lineage potentials [Pina et al., 2008]. Nevertheless, mechanisms by which HSC commit to and differentiate down the different blood lineages are still debated [Pina et al., 2008].
Human erythroid differentiation-associated Gene (EDAG) (also known hemgn in mouse, and RP59 in rat) is a hematopoietic tissue-specific gene and involved in the regulation of proliferation, differentiation and apoptosis of hematopoietic cells [Li et al., 2004]. Several previous studies have examined the gene expression patterns and effect of ectopic EDAG expression on cellular phenotype. Yang et al. 2001 reported that EDAG is primarily expressed in the linloc-kit+Sca-1+ hematopoietic stem cell population and CD34+ progenitor cells, and is down-regulated in mature blood cells. An et al. 2005 found that EDAG is overexpressed in PBMCs of patients with leukemia, and the association of EDAG overexpression and poor prognosis in de novo acute myeloid leukemia (AML) has also been identified, suggesting that EDAG may play a modulator role in AML. Lu et al. 2001 reported that EDAG is down-regulated during erythroid or megakaryocytic differentiation of human erythroleukemia cell line K562 cells by treatment with hemin, erythropoietin or PMA. Recently, EDAG was identified as a novel direct target of transcription factor GATA-1, and PMA-mediated inhibition of the EDAG gene is mediated through down-regulation of transcription factor GATA-1 and involved PKC/MAPK signaling pathway [Yang et al., 2006; Li et al., 2008]. Li et al. 2004 reported that down-regulation of EDAG expression using antisense in K562 cells results in inhibition of cell growth and colony formation, and enhancement of sensitivity to erythroid differentiation induced by hemin. Overexpression of EDAG in HL-60 cells significantly blocked the expression of the monocyte/macrophage differentiation marker CD11b after pentahydroxytiglia myristate acetate induction [Li et al., 2004]. Moreover, enforced expression of EDAG in pro-B Ba/F3 cells prolonged survival and increased the expression of c-Myc, Bcl-2 and Bcl-xL in the absence of interleukin-3 (IL-3) [Li et al., 2004]. Overexpression of EDAG in hematopoietic cells suppresses the lymphoid lineage development while enhances myeloid development in transgenic mice [Li et al., 2007]. However, the molecular mechanism of EDAG in hematopoietic cell differentiation has been much less documented.
In the present study, we provided convincing evidence that introduction of an EDAG expression vector into IL-3-dependent murine myeloid cell line 32D cells promoted differentiation along the erythroid/megakaryocytic lineage. We also found that several erythroid transcription factor GATA-1 and its target genes were up-regulated in EDAG overexpressed 32D cells. Moreover, knockdown of EDAG protein in K562 cells resulted in the decreased mRNA expression of these genes. These results suggested that EDAG might function as a positive regulator of erythroid/megakaryocytic differentiation associated with the induction of GATA-1 and its target genes.
MATERIALS AND METHODS
Cell Culture and Stable Transfection
Human erythroleukemia cell line K562 was obtained from the National Institute of Biological Products (Beijing, China) and maintained in RPMI 1640 medium with 10% heat-inactivated FCS, 2 mM glutamine, 100 IU/ml penicillin, 100 mg/ml streptomycin, 2 g/L sodium bicarbonate and 10 mM HEPES in a 37°C incubator with 5% CO2. The IL-3 dependent murine myeloid cell line 32D was purchased from the American Type Culture Collection (Rockville, MD) and cultured in RPMI 1640 (Gibco Invitrogen, CA) supplemented with 10% WEHI-conditioned medium as a source of IL-3, 10% fetal calf serum at 37°C with 5% CO2. For stable transfection, cells (5 × 106) were washed, resuspended in 0.5 ml of ice-cold phosphate-buffered saline (PBS), mixed with linearized pcDNA3.1 (His/Myc)-EDAG [Li et al., 2004] or control plasmid DNA (15 µg) and subjected to electroporation (960 mF; 220 V) in a 0.4-cm-wide electroporation cuvette. Cells were then cultured in 10 ml of complete RPMI 1640 medium for 2 days and selected with 800 mg/ml G418. Monoclones of the transfectants were obtained using the limiting dilution method and identified by Western blotting.
MRNA Microarray Analysis
Stably transfected cells and normal 32D cells were harvest, and total RNA was extracted with Trizol Reagent (BD Biosciences Clontech, Palo Alto, CA) and ethanol precipitation. The oligo dT-primed fraction of total RNA (60 µg) was used to synthesize labeled cDNA probes in the presence of either Cy3-dUTP or Cy5-dUTP and SuperScript II reverse transcriptase (Invitrogen). Labeled cDNA was hybridized to 36k Mouse Genome Array Genechips (CapitalBio, Beijing, China) which contains probe sets for about 25,000 murine genes. Array hybridizations and subsequent scanning, visualization and quantitation were performed. After background correction and removal of bad spots, a space and intensity-dependent normalization based on a LOWESS program was employed. For each test and control sample, two hybridizations were performed by using a reversal fluorescent strategy, the geometric mean of two dye-swap slides was calculated as final ratio. The most up- and down-regulated genes associated with EDAG expression were screened for biological process and cellular component gene ontology (GO) annotations using the Gene Ontology database (http://biorag.org/index.php). All data is MIAME compliant. Microarray data reported herein have been deposited at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE18862.
Reverse Transcription and Real-Time PCR
Total RNA isolation and reverse-transcription were applied according to the manufacturer's protocol. The cDNA was analyzed using real-time PCR according to the instruction from the kit. In brief, real-time PCR was done using Bio-Rad IQTM5 Multicolor Real-time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) and SYBR Premix Ex Taq™ (2×) kit (TaKaRa, Japan). The cycling conditions were as follows: 95°C for 1 min, 40 cycles of 10 s at 95°C, 30 s at 60°C and 30 s at 72°C. SYBR Green fluorescence was measured after each elongation step. Specific primers for each gene were listed in Supplementary file 2. At the end of PCR, a melting curve analysis was performed by gradually increasing the temperature from 55 to 95°C to determine purity. PCR was set up in triplicates and threshold cycle (Ct) values of the target genes were normalized to the endogenous control. Differential expression was calculated according to the 2−ΔΔCt method. All samples were independently analyzed at least twice for each gene. The housekeeping gene GAPDH served as an additional control for the cDNA quality.
Flow Cytometric Analysis
Cells (2 × 105) were pelleted at 700g for 5 min and washed with PBS containing 2% human albumin (PBS-AB). The cells were resuspended in 100 µl PBS-AB with 2 µl human CD61 monoclonal antibody (11-0611, eBioscience) conjugated with fluorescein isothiocyanate (FITC), incubated at 4°C for 20 min and washed twice with PBS-AB. The cells were resuspended in 400 ml PBS-AB and were examined by flow cytometry using a FACSCAN system (Becton Dickinson).
For cell cycle analysis, cells were fixed with ice-cold absolute ethanol or with 0.5% paraformaldehyde for 15 min before with absolute ethanol, washed with PBS, and resuspended in 0.2 ml of RNase A (1 mg/ml) for 30 min at 37°C. After being stained with 0.3 ml propidium iodide (60 µg/ml), the cells (1 × 105) were analyzed for DNA content with a flow cytometer (Becton Dickinson).
Western Blotting
For Western blotting, cells were lysed with M-PER® Mammalian Protein Extraction Reagent (Pierce, Rockford, IL). Then, Western blot analysis was performed according to standard procedures. Antibodies were used at the following concentrations: EDAG antiserum, 1:1,000; GATA-1 (sc-266, Santa Cruz, CA), 1:1000; PU.1 (sc-352, Santa Cruz), 1:1000; β-actin (sc-47778, Santa Cruz), 1:1,000. Chemiluminescent detection was conducted using supersignal substrate (Pierce) according to the manufacturer's specifications.
RNA Interference (RNAi)
The small interfering RNA (siRNA) oligos of EDAG were synthesized in GenePharma Biotechnology, the sequences are as follows: siEDAG-1: sense: 5′-CCU GAA ACG UAU CAA GAA ATT-3′; antisense: 5′-UUU CUU GAU ACG UUU CAG GTT-3′); siEDAG-2: sense: 5′-AUA AGG AUG UGC CUA AAG A-3′; antisense: 5′-UCU UUA GGC ACA UCC UUA U-3′. The negative control (nonsilencing) siRNA (GenePharma, Shanghai, China) were used in control experiments. RNAs were transfected into K562 cells by electroporation (975 µF; 135V in a 0.2-cm-wide electroporation cuvette) at a concentration of 1 µM.
Luciferase Assay
Cells were plated in 6-well tissue culture plate at a density of 2 × 105 cell per well, and transfected with 2 µg of each construct reporter plasmid using electroporation. As an internal control, the pRL-TK vector was cotransfected which led to the constitutive expression of Rennila luciferase (Promega Corp., Madison, WI). Twenty-four hours after transfection, cells were harvested in passive lysis buffer, and activities of firefly and Rennila luciferase were measured with the Dual Luciferase Assay system (Promega) according to the manufacturer's instructions. The activity of firefly luciferase was normalized by the activity of Rennila luciferase. All transfection and reporter assays were performed independently at least three times.
Benzidine Staining
Cells were washed twice with ice-cold PBS and then resuspended in ice-cold PBS. The benzidine solution (1 ml), which contained 0.2% DMBZ (3,3′-dimethoxybenzidine) (Sigma–Aldrich, St. Louis, MO) and hydrogen peroxide (final concentration 0.0012%), was added and incubated for 10 min at room temperature. Benzidine-positive cells were examined and quantitated under the microscope. At least 100 cells were counted in triplicate for each condition.
Eosinophil Peroxidase Activity Assay
1 × 106 cells were collected and washed twice with ice-cold PBS. Eosinophil peroxidase (EPX) was solubilized from cells in PBS by sonication. After centrifugation at 12,000 rpm for 15 min at 4°C, the supernatant was used to measure EPX activity with rabbit IgG-HRP (sc-2004, Santa Cruz) as positive control. EPX activity was measured by the o-phenylenediamine (OPD) method as previously reported [Kroegel et al., 1989]. Briefly, OPD (Sigma–Aldrich) substrate solution is prepared with 0.1 mM OPD in a 0.05 M Tris-Buffer (pH 8.0) containing 0.1% Triton X-100 and 1 mM hydrogen peroxide. The OPD was made up as a 10 mM stock solution in water, stored at −80°C and diluted immediately before use. 300 µl of this substrate were added to 150 µl of the samples and incubated for 30 min at 37°C. Reaction was then stopped with 200 µl of 4 M H2SO4. The absorbance was then determined at 492 nm by using spectrophotometer (Bio-Rad).
Statistical Analysis
All experiments were performed at least three times. Data were reported as means ± SEM and the statistical analysis was performed using the Student's t-test. A value of P ≤ 0.05 was considered to be significant.
RESULTS
Overexpression of EDAG Induces Erythroid/Megakaryocytic Differentiation in IL-3-Dependent 32D Cells
To detect whether overexpression of EDAG changed the lineage phenotype of hematopoietic cells, IL-3-dependent 32D cells were transfected with pcDNA3.1 (His/Myc)-EDAG or the empty vector, and the stably transfected cell clones were selected using G418. Expression of EDAG was verified by Western blot analysis and real-time PCR (Fig. 1). As shown in Figure 1A, EDAG protein was hardly detected in a parental 32D cells or control 32D cells transfected with empty vector, but it was highly expressed in EDAG transfectants with the expression level similar to that in a human erythroleukemia cell line K562. Two monoclones with different expression level of EDAG named 32D/EDAG-1 and 2 were identified.
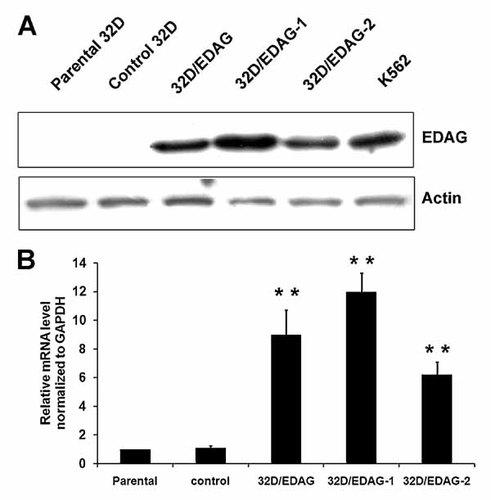
Detection of exogenous expression of EDAG in 32D cells. 32D cells were transfected with an o EDAG expression vector pcDNA3.1-EDAG or a control vector, and the stably transfected cell clones were selected using G418 for 2 weeks. Then cell lysates were extracted for Western blotting (A) and β-actin was used as internal control. Total RNA were extracted for Real-time PCR (B). Results were expressed as fold induction compared with cells transfected with control cells and normalized to GAPDH mRNA. Each bar represented the mean ± SD for three independent experiments. The statistical difference between the samples was demonstrated as *P ≤ 0.05 or **P ≤ 0.001.
Possible effects of the ectopic expression of EDAG on the growth of 32D cells were first investigated. Control 32D cells, 32D/EDAG-1, and 32D/EDAG-2 cells were cultured in the presence or absence of cytokines IL-3 and cell proliferation was measured. No significant difference was observed in the growth kinetics of all these cells in the presence of IL-3 (Fig. 2A). Although no cytokine-independent growth was observed in any of the cell lines, the growth of 32D/EDAG cells was markedly reduced in absence of IL-3 (Fig. 2B). After IL-3 was withdrawn for 24 h, control cells still contained some live cells, and the average viability was 72.89%, whereas that of 32D-EDAGs cells was only 7.50% and 4.65%, respectively. Moreover, significant reduction of 32D/EDAG cells growth was also detected in the presence of EPO, G-CSF or GM-CSF (Fig. 2C–E). Apoptosis was further investigated. Just as Figure 2F shows, under IL-3 starvation, all the cell lines extensively underwent G0/G1 arrest (64.87, 74.07 and 78.45%, respectively) after 24 h and apoptosis, however, 32D/EDAG-1 cells showed more than 21% of cells underwent apoptosis and concomitant with an decrease in the G2-M phase (from 21.16% to 5.61%). The similar results were obtained in 32D/EDAG-2 cells. These data suggested that enforced expression of EDAG inhibited cell growth and enhanced cell apoptotic after IL-3 was withdrawn.
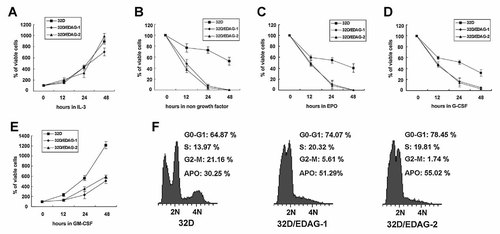
Overexpression of EDAG suppresses cell growth and causes rapid cell apoptosis in the absence of IL-3. 32D, 32D/EDAG-1, 32D/EDAG-2 cells were cultured in the presence of IL-3 (A) or in the absence of IL-3 (B) or in the presence of EPO (C), G-CSF (D), or GM-CSF (E) for the time indicated and then the cell growth was determined by MTS assay. F: Cells were cultured in the absence of IL-3 for 24 h then the cells were harvested for cell cycle analysis by a FACSCAN system.
Previous studies suggest that in 32D cells terminal differentiation might attribute to the increased cell apoptosis and cell cycle arrest. Then we detected the effect of EDAG on the differentiation of 32D cells. We first investigated the expression of β-globin, a marker of mature erythroid cells, by benzidine staining. Although control 32D cells were essentially negative on benzidine staining, approximately 25% of 32D/EDAG cells, 40% of 32D/EDAG-1 and 18% of 32D/EDAG-2 cells became positive (Fig. 3A,B). Also, the erythroid cells specific surface marker TER119 was investigated. As shown in Figure 3C, TER119 positive cells were scarcely detected in 32D cells, whereas an easily detectable level of TER119 positive cells (6.2%) was observed in EDAG transfectants. Next, we examined the megakaryocytic surface expression of lineage-specific antigens CD61 (GP IIIa) and the result suggested that the number of CD61 positive cells was increased from 7.1% to 23% with EDAG overexpression. Moreover, the megakaryocytic characteristics of these clones were confirmed by Wright-Giemsa staining (data not shown). Furthermore, reduced expression level of the myeloid marker CD11b and monocytes marker F4/80 were detected in 32D/EDAG cells (Fig. 3C). These results suggested that the ectopic expression of EDAG reprogrammed 32D cells toward an erythrocytic/megakaryocytic phenotype.
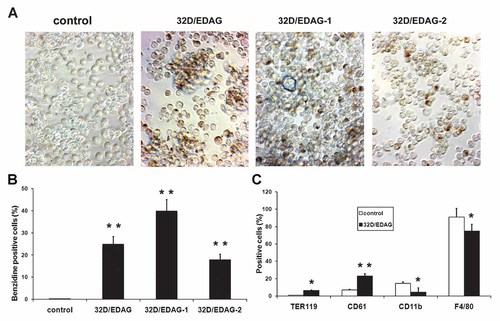
Overexpression of EDAG induces erythroid/megakaryocytic differentiation in 32D cells. A: 32D/EDAG clones and control 32D cells were stained with benzidine and benzidine-positive cells were counted. Quantitation of benzidine-positive cells was described in (B). C: Cells were incubated with the antibodies indicated and analyzed by flow cytometry analysis. Each bar represented the mean ± SD for three independent experiments. The statistical difference between the samples was demonstrated as *P ≤ 0.05 or **P ≤ 0.001. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Microarray Analysis Identified Genes Differentially Expressed as a Result of Enforced EDAG Expression
To elucidate the mechanism of differentiation induction by enforced EDAG, we compared the gene expression profiles of cells with EDAG overexpression by microarray analysis. Total RNA from the control 32D cells and 32D/EDAG cells were used to generate target cRNA and hybridized to 36k Mouse Genome Array Genechips, representing, around 25,000 characterized murine genes (GEO accession number GSE18862). For data analysis, arrays were normalized to a set of housekeeping genes, and expression profiles of EDAG enforced-expressing clones were compared to mock transfected clones, using the latter as baseline. Using a 2-fold change cut-off, we identified 332 gene probes with decreased expression and 288 gene probes with increased expression in response to the overexpression of EDAG in 32D cells (Supplementary File 1).
The list of down-regulated genes included genes encoding specific markers of myeloid, lymphoid and mast cells (Table I). These genes included CD14, CD52, CD137 and GP49a. Most interestingly, many of the up-regulated genes were hematopoietic transcription factors which are regulated by the GATA-1 (EKLF, NF-E2, Gfi-1B, hemogen, etc.) and play key roles in erythropoiesis and megakaryocytic development (GATA-1, SCL, Evi-1, etc.). Furthermore, the erythroid marker hemoglobin α and β were increased significantly. Also the megakaryocytic specific gene GPIX was up-regulated. In addition, eosinophil specific-expression genes eosinophil peroxidase (EPX) precursor and eosinophil granule major basic protein also were significantly increased by 599 folds and 26 folds, respectively.
| Gene name | CBC. annotation | Fold change | Description |
|---|---|---|---|
| GATA-1 target genes | |||
| Epx | NM_007946 | 599.239 | Eosinophil peroxidase precursor |
| Hbb | XM_489729 | 176.553 | Predicted: hemoglobin beta chain complex |
| Hbb-b2 | NM_016956 | 51.123 | Hemoglobin, beta adult minor chain |
| Prg2 | NM_008920 | 26.659 | Eosinophil granule major basic protein precursor(MBP) |
| Gata1 | NM_008089 | 24.677 | GATA binding protein 1 (Gata1) |
| Hba-a1 | NM_008218 | 23.467 | Hemoglobin alpha, adult chain 1 |
| Hbb-y | NM_008221 | 17.943 | Hemoglobin Y, beta-like embryonic chain |
| Hbb-b1 | NM_008220 | 15.345 | Hemoglobin, beta adult major chain |
| Hba-x | NM_010405 | 12.163 | Hemoglobin X, alpha-like embryonic chain in Hba complex |
| Evi1 | NM_007963 | 7.987 | Ecotropic viral integration site 1 |
| Klf1 | NM_010635 | 3.274 | Erythroid krueppel-like transcription factor (EKLF) |
| Gfi1b | NM_008114 | 3.146 | Growth factor independent 1B |
| Nfe2 | NM_008685 | 2.279 | Nuclear factor, erythroid derived 2 |
| Hemgn | NM_053149 | 2.694 | Hemogen (mouse) |
| TAL-1(scl) | NM_011527 | 2.507 | T-cell acute lymphocytic leukemia 1 (Tal1) |
| GPIX | NM_018762 | 6.935 | Glycoprotein 9 (platelet); platelet glycoprotein IX |
| Surface marker | |||
| CD14 | NM_009841 | 0.0117 | Monocyte differentiation antigen CD14 precursor |
| CD52 | NM_013706 | 0.0086 | Lymphocyte differentiation antigen B7 |
| CD137 | NM_011612 | 0.1601 | T-cell antigen 4-1BB (CD137 antigen) |
| GP49A | NM_008147 | 0.0136 | Mast cell surface glycoprotein GP49A precursor |
| MPO | NM_010824 | 0.378 | Myeloperoxidase precursor (EC 1.11.1.7) (MPO) |
In addition to hematopoietic differentiation related genes, a series of genes with diverse biologic roles were also identified as target genes down-regulated or up-regulated by EDAG. These genes are involved in cell-cycle control, apoptosis, transcriptional regulation cell signal transduction (Supplementary File 1).
Validation of Up-Regulation of Candidate Target Genes in Response to Enforced EDAG Expression
To validate expression levels by an independent and more quantitative method, we employed real-time quantitative RT-PCR to confirm the results obtained by microarray analysis for the 15 candidate target genes including several erythroid transcription factors and GATA-1 target genes. As Figure 4A shows, the data obtained by microarray analysis were verified by this independent approach, indicating that our transcriptional profiling accurately reflected gene expression levels in 32D/EDAG cells.
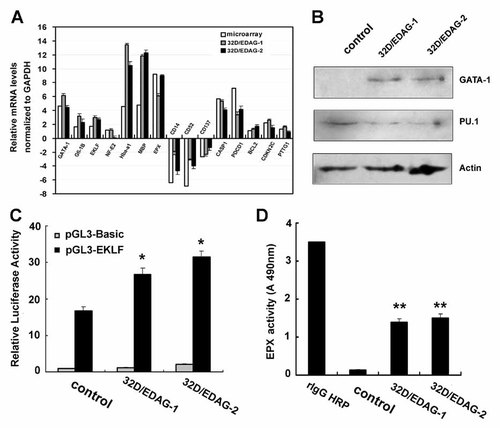
Validation of selected genes regulated by EDAG in 32D cells. A: 32D cells, 32D/EDAG-1, and 32D/EDAG-2 cells were harvested and total RNA were extracted for real-time PCR analysis of the indicated gene expression. Result were presented as fold induction compared with control cells and normalized to GAPDH mRNA. The fold change of the microarray was shown as “Microarray.” B: Total cell lysates were prepared for Western blotting with the antibodies as indicated. β-Actin was used as internal control. C: 32D cells, 32D/EDAG-1 cells and 32D/EDAG-2 cells were transfected with a EKLF promoter reporter vector (pGL3-EKLF) or pGL3-Basic control vector for 24 h using TK as a internal control. Then cell were harvested for luciferase activity assay. D: EPX activity assay was performed, using rabbit IgG-HRP as positive control. Results represented mean ± SD of three independent experiments. The statistical difference between the samples was demonstrated as *P ≤ 0.05 or **P ≤ 0.001.
Since previous studies have shown that the development of the erythroid and myeloid blood cell lineages is controlled in part by the master transcriptional regulators GATA-1 and PU.1 [Nerlov et al., 2000]. The two proteins have been shown to function in an antagonistic fashion, with GATA-1 repressing PU.1 activity during erythropoiesis and PU.1 repressing GATA-1 function during myelopoiesis [Nerlov et al., 2000]. Therefore, we further confirm the expression of GATA-1 and PU.1 by Western blot analysis. GATA-1 was up-regulated while PU.1 was slightly down-regulated in 32D/EDAG-1 and 32D/EDAG-2 cells compare with control transfected cells (Fig. 4B).
We further confirm the up-regulation of EKLF which is a transcriptional target of GATA-1, we analyzed the activity of promoter of EKLF which contains GATA-1 binding sites in 32D cells and 32D/EDAG cells by luciferase activity assay. The EKLF promoter construct was transfected into 32D/EDAG cells or 32D/Mock cells with a Rennila luciferase plasmid as an internal transfection control. The activity of EKLF promoter was increased up to 160% in 32D/EDAG-1 and 190% in 32D/EDAG-2 cells compared with 32D/Mock cells (Fig. 4C).
EPX is an eosinophil specific-expression genes. To confirm the up-regulation expression of the eosinophil peroxidase gene was correlated with the increased functional level, enzyme activity of EPX was assayed. Just as shown in Figure 4D, the enzyme activity of EPX was increased in 32D/EDAG cells compare with control cells.
Silencing of Endogenous EDAG in K562 Cells Leads to Altered Expression of GATA-1 and Its Target Genes
Since enforced expression of EDAG up-regulated GATA-1 and its target genes in 32D cells, we performed RNA interference assay to investigate whether knockdown of EDAG endogenous expression altered the expression of these genes in physiological conditions. K562 cells, in which EDAG was highly expressed, were transfected with siRNA directed against EDAG (siEDAG-1) or control siRNA. As shown in Figure 5, transfection of K562 cells with EDAG siRNA resulted in an up to 88% decrease in the EDAG mRNA level (Fig. 5B) and a 73% decrease in the protein level (Fig. 5A) within 48 h after transfection. However, transfection with non-specific RNA (NC) had no effect on EDAG protein and mRNA level (data not shown). Furthermore, the levels of β-actin were not affected by non-specific siRNAs (data not shown). When endogenous EDAG expression was knocked down by siRNA, K562 cells showed an decreased expression of GATA-1, NF-E2, Gfi-1b, HBB, EKLF and EPX genes. The reduced protein level of GATA-1 was confirmed by Western blotting analysis. To rule out off-target effects, another siRNA oligo against EDAG was used and the similar result was obtained (data not shown). These results indicated that EDAG might play a physical role in the regulation of expression of GATA-1 in K562 cells.
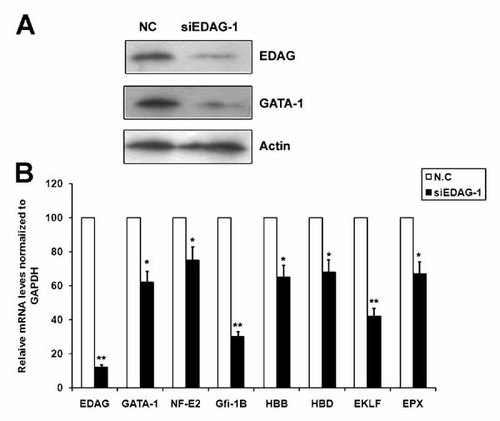
Knockdown of EDAG decreases the expression of GATA-1 and its target genes in K562 cells. K562 cells were transfected with EDAG siRNA (siEDAG-1) or control siRNA duplex (NC) for 48 h. Total cell lysates were prepared for Western blotting with β-actin as the internal control (A) and total RNA were extracted for real-time PCR with GAPDH as the internal control (B). Results represented mean ± SD of three independent experiments. The statistical difference between the samples was demonstrated as *P ≤ 0.05 or **P ≤ 0.001.
DISCUSSION
32D is a diploid interleukin-3 (IL-3) dependent cell line established from long term marrow cultures of C3He/J mice injected with the Friend murine leukemia virus [Migliaccio et al., 1989]. This cell line, in particular, generates at low frequency cells that respond to EPO, GM-CSF, or G-CSF and that differentiate in the presence of these growth factors along the appropriate hematopoietic lineage [Migliaccio et al., 1989]. Therefore, although the majority of these cells have a mast cell morphology, it has been used to study the process of control of normal hematopoiesis [Migliaccio et al., 1989]. EDAG, was originally cloned as an erythroid differentiation regulatory gene [Yang et al., 2001]. In fact, EDAG, as a GATA1 target gene, is expressed in HSCs, HPCs and CD71+ cells in bone marrow [Yang et al., 2006; Ceredig et al., 2009]. Its down-regulation during the differentiation of HSCs is critical for the lymphoid development [Li et al., 2007]. Increased EDAG expression promotes myeloid-cell development and blocks B- and T-cell development [Li et al., 2007]. These observations prompted investigation of the effects of the levels of EDAG expression on lineage commitment. In the present study, we found that the overexpression of EDAG in 32D cells leads to erythroid and megakaryocytic differentiation. Erythroid cells were confirmed by expression of hemoglobin and erythroid specific surface marker TER119. Megakaryocytes were verified by the morphology and the expression of megakaryocytic surface antigens CD61. This finding indicated that the EDAG can function as a positive regulator of erythroid/megakaryocytic differentiation. In addition, we also found that 32D/EDAG cells expressed high level of EPX activity, suggesting a role of EDAG in regulation of eosinophil differentiation.
The molecular mechanism of EDAG regulating hematopoiesis differentiation is remained to be determined. In this article, we employed microarray analysis to identify potential downstream targets of EDAG, with the aim of detecting the pathways regulated by EDAG. This approach revealed that a large number of genes were differentially expressed in response to the enforced expression of EDAG. Focusing on genes showing increased or decreased levels of expression and using a twofold change cutoff, we identified 332 genes with reduced expression and 288 genes with increased expression. Interestingly, among up-regulation genes, we find that EDAG-overexpressing 32D cells displayed higher expression of erythroid transcription factor GATA-1 and its target genes including erythroid transcription factors EKLF, NF-E2, Gfi-1b, hemogen, SCL and erythroid specific protein hemoglobin. These data was consistent with the erythroid phenotype of 32D/EDAG cells with increased number of benzidine-positive cells and up-regulation of erythroid specific gene TER119. Furthermore, the megakaryocytic specific gene GPIX which contains a GATA-binding motif in its promoter [Bastian et al., 1996] was also increased and the megakaryocytic marker CD61 was induced significantly, suggesting a positive role of EDAG in megakaryocytic differentiation. Among down-regulation genes, myeloid marker CD11b, mast cell marker GP49A, and monocyte marker CD14 were found. Also, the expression level of the myeloid transcription factor PU.1 was decreased. These results indicated that EDAG affects hematopoietic differentiation by altering the expression level of genes involved in different lineage commitment.
The expression or activities of lineage-restricted factors and/or in particular lineage-restricted transcription factor(s) may be a key step in regulating hematopoietic cell differentiation. GATA-1 is a hematopoietic transcription factor that is expressed in erythroid, megakaryocytic, eosinophil and mast cell precursors [Cantor and Orkin, 2002; Ferreira et al., 2005]. Several previous studies have examined the effect of ectopic GATA-1 expression on cellular phenotype and gene expression patterns in different hematopoietic cell lines such as transformed avian myeloblastic cell lines, murine myeloid FDCP2 cell line, myeloid leukemic cell M1 line, and early myeloid cell line 416B [Layon et al., 2007]. For example, Kulessa et al. 1995 found that overexpression of GATA-1 in transformed avian myeloblastic cell lines under various conditions can lead to phenotypic conversion to erythroid, eosinophilic or megakaryocytic cell types. Forced GATA-1 expression in the M1 murine myeloid leukemia cell line resulted in the formation of mixtures of megakaryocytic and erythroid cells within individual clones and activation of erythroid and megakaryocytic genes [Yamaguchi et al., 1998]. Given the importance and association of GATA-1 with the erythroid and megakaryocytic differentiation, and overexpression of EDAG shared similar cellular phenotype with that of GATA-1, a more detailed analysis of EDAG functional relationship with GATA-1 is studied. We confirmed the up-regulation of GATA-1 by Western blotting. Furthermore, the increased expression of the GATA-1 correlated with the increased functional activity since one of GATA-1 target genes EKLF showed increased promoter activity in 32D/EDAG cells. Also the hemoglobin proteins which are GATA-1 target genes including hemoglobin-α and β were also up-regulated. Interestingly, the mouse homolog of EDAG in mouse named Hemgn, which is also a target gene of GATA-1 was also increased. Moreover, our previous study found that the specific expression of EDAG in erythroid and megakaryocytic precursor cells of adult bone marrow coincides with the expression profile of GATA-1. Furthermore, we found that knockdown of endogenous EDAG expression by siRNA resulted in decreased expression of GATA-1 and its target genes in K562 cells. These results indicated that up-regulation of GATA-1 and in turn activation of the transcription GATA-1 target genes may play an important role in regulation of erythroid/megakaryocytic differentiation of enforced EDAG expression in 32D cells.
Our previous study suggest that down-regulation of EDAG in K562 cells by antisense oligonucleotides leads to a increased erythroid differentiation sensitivity in response to Hemin [Li et al., 2004]. In the present study, we showed that overexpression of EDAG in 32D cells also induced erythroid differentiation. The similar phenotypes observed after EDAG overexpression or silencing through antisense oligonucleotides may be due to several reasons: First, K562 cells is a erythroleukemia cell line, while 32D is considered to be a population of immortalized “normal” hematopoietic progenitors. Leukemic cell lines do not necessarily mimic corresponding normal hematopoietic cells. Furthermore, differences in the EDAG expression level should be considered. There may be a window of appropriate EDAG expression levels for erythroid differentiation. One could imagine that overexpression of EDAG may inhibit the function of endogenous EDAG protein complexes by disrupting the normal stoichiometry of these complexes. In fact, similar observations have been made in other genes. Silencing or overexpression of the Notch ligand DLL4 in ECs led to a significant inhibition of proliferation, and overexpression or knock-out of Notch4 produced similar vascular phenotypes in mice [Iso et al., 2003; Patel et al., 2005; Williams et al., 2006]. THAP1 overexpression or silencing also inhibits the cell proliferation and cell-cycle progression [Cayrol et al., 2007]. Furthermore, the hematopoietic transcription factor SCL shows tremendous similarity with EDAG. Overexpression in HSCs stimulates primitive erythroid, and megakaryocytic differentiation while antisense of SCL specifically inhibits proliferation and self-renewal of K562 cells with enhancing their spontaneous erythroid differentiation[Green et al., 1991; Valtieri et al., 1998]. In the present study, we found that in 32D/EDAG cells, the expression level of SCL were also increased, which may suggest a role of SCL in the erythroid differentiation induced by EDAG.
Our previous study suggests that overexpression of EDAG in a pro-B cell line Ba/F3 protects cells from apoptosis through the activation of NF-Kb [Li et al., 2004]. Also, EDAG leads to increased levels of c-Myc, Bcl-2 and Bcl-xL [Li et al., 2004]. Moreover, overexpression of EDAG in NIH3T3 cells causes malignant transformation [Lu et al., 2002]. However, in this study, overexpression of EDAG in 32D cells suppresses cell growth in the presence of GM-CSF, EPO, or G-CSF and provokes rapid apoptosis in the absence of IL-3. It may be due to the highly induced expression of GATA-1 and hemoglobin, and the effect of altered differentiation of 32D cells to a erythroid phenotype. It was reported that GATA-1 promotes both erythroid maturation and G1 cell cycle arrest of G1E cells [Rylski et al., 2003]. Molecular studies combined with microarray transcriptome analysis revealed an extensive GATA-1-regulated program of cell cycle control in which numerous growth inhibitors were upregulated and mitogenic genes were repressed [Rylski et al., 2003]. GATA-1 inhibited expression of cyclin-dependent kinase (Cdk) 6 and cyclin D2 and induced the Cdk inhibitors p18INK4C and p27Kip1 with associated inactivation of all G1 Cdks [Rylski et al., 2003]. Upregulation of alpha globin promotes apoptotic cell death in the hematopoietic cell line FL5.12 [Brecht et al., 2005]. Enrichment of FL5.12 cells expressing GFP-alpha globin was difficult even in the presence of IL-3. Caspase-8, -9 and -3 as well as the proapoptotic factor Bax and cytochrome c were activated [Brecht et al., 2005]. Our microarray analysis suggested that the Cdk 5 was reduced while p18INK4C was induced significantly in 32D-EDAG cells which is similar to that in GATA-1 overexpressing cells. Although the caspase 1, 4, and 12 were induced in 32D-EDAG cells, the anti-apoptotic factor Bcl-2 was induced to about twofold, suggesting a more complex mechanism of EDAG in regulating the cell cycle and apoptosis.
In conclusion, forced expression of EDAG in the myeloid cell line, 32D, induces the differentiation towards erythroid and megakaryocytic lineage associated with the induction of GATA-1 and its target genes.