Expression profiles of SV40-immortalization-associated genes upregulated in various human cancers
Abstract
Immortalization is an early and essential step of human carcinogenesis which is associated with alterations in gene expression and regulation. Suppression subtractive hybridization (SSH) was successfully performed to identify immortalization-associated genes upregulated in SV40-immortalized lung fibroblasts. We identified 116 known genes which were related to diverse functions, with 32.8% relevant for cell cycle or proliferation indicating the potential involvement of these genes in immortalization. We chose eight known genes located on the overrepresented chromosomes of non-small-cell lung cancers (NSCLCs). ASPM, RFC4, C3orf26, BXDC2, C15orf44, AURKA, C20orf77, and RBMX were upregulated in immortalized cells, cancer cells, and non-small-cell lung cancer (NSCLC) tissues. We additionally cloned two novel genes (CHA-V-97 and CHA-V-165) which showed similar upregulated expression patterns in cells and tissues examined. Identification and further characterization of these genes may provide insights of novel players for immortalization and human carcinogenesis. J. Cell. Biochem. 106: 703–713, 2009. © 2009 Wiley-Liss, Inc.
Immortality is an essential characteristic for the development of cancers. Genetic alterations underlying cell immortalization involved in carcinogenesis remain to be understood. There are numerous studies in cell culture systems, in which genetic alteration by simian virus 40 (SV40) T antigen [Ozer et al., 1996], human papillomavirus type 16 (HPV-16) E6 or E7 genes [Shiga et al., 1997], Bmi-1 oncogene [Dimri et al., 2002], and human telomerase reverse transcriptase (hTERT) [Bodnar et al., 1998] can immortalize somatic cells. Repeated subculture of normal human somatic cells in culture, exposure to oxidative stress, or activation of oncogenes can cause cells to gradually reduce their proliferation rate and enter replicative senescence, an irreversible growth arrest, accompanied with continued metabolic activity [Lundberg et al., 2000]. Immortalization of cells can be induced by overcoming the following crisis stages of replicative senescence: mortality stage 1 (M1) leads to mitogen incapacity and bypasses the cellular DNA replication activity of the T antigen, which arrests cells near at the G1/S interface; cell division subsequently malfunctions during mortality stage 2 (M2) [Wright et al., 1989]. During the crisis period, cell proliferation and cell death are balanced.
Because appropriate cellular growth and proliferation involve many physiological processes, several levels of regulatory machinery govern the operation of the cell cycle: receptor–ligand interactions at the cell surface, intracellular signal transduction cascades, transcriptional networks, and control circuits [Hahn and Weinberg, 2002]. Previous reports have demonstrated various regulatory proteins involved in p53 and p16/Rb pathways as a potential mediator for cellular senescence. The most prominent candidate is p16INK4A, whose expression gradually increases as fibroblasts approach senescence [Alcorta et al., 1996]. In human keratinocytes, inactivation of p16INK4A is required to activate telomerase expression [Dickson et al., 2000]. Another senescence-inducing signal, p19ARF, directly binds to and inhibits the function of MDM2 (Mdm2 p53 binding protein homolog), the factor that induces degradation of the p53 tumor suppressor protein. Malfunction of MDM2 results in p53-mediated growth arrest [Sherr and Weber, 2000]. Blockage of such signaling pathways allows cells to immortalize by escaping the restrictions placed on their division capacity. Although we currently possess some understanding of many of the regulatory pathways involved in cellular proliferation, the mechanisms underlying the immortalization process remains unclear.
Lung cancer is one of the most common and deadly forms of human cancer with an estimated 213,380 new cases and 160,390 deaths in the USA anticipated for 2007. Moreover, the death rate is continuously increasing both in men and in women [Jemal et al., 2007]. Lung cancer is classified into either small-cell lung carcinoma (SCLC) (20% of all) or non-small-cell lung carcinoma (NSCLC) (80% of all). NSCLC is further divided into adenocarcinoma (AC) (30–35%), squamous cell carcinoma (SCC) (30%), large cell carcinoma (LCC) (10%), and NSCLC subtypes, such as adenosquamous cell carcinoma (ASCC) and bronchioalveolar carcinoma (BAC). The pathogenesis of lung cancer still remains indefinable due to its aggressive biological nature and heterogeneity. Many genes are known to be involved in carcinogenesis as well as the genesis of NSCLC. The tumor suppressor gene p53 [Han et al., 2002], the vascular endothelial growth factor receptor (VEGFR) [Liao et al., 2001], the proliferation factor Ki-67 [Shiba et al., 2000], and the anti-apoptotic factor Bcl-2 [Martin et al., 2003] are potentially involved in NSCLC. However, none of these genes have been used in diagnosis or prognosis of NSCLC. Development of lung cancer is associated with abundant molecular changes including many genes which are currently unknown.
We used suppression subtractive hybridization (SSH) method in order to identify genes that are totally unknown. In this study, we found two novel genes that were strongly upregulated in immortal cells compared to senescent cells. These two novel genes were also upregulated in diverse cancer cells and NSCLCs. We also screened and identified known genes that are upregulated in SV40-mediated immortalized fibroblasts (WI-38 VA13) but not in equivalent senescent cells (WI-38). We also selected eight known genes and investigated their expression patterns in various cancer cell lines and immortalized cell lines, suggesting the potential involvement of these genes in NSCLCs.
Our study provides novel information that known genes (ASPM, RFC4, C3orf26, BXDC2, C15orf44, AURKA, C20orf77, and RBMX) and novel genes (CHA-V-97 and CHA-V-165) may be involved in the unknown mechanisms underlying the immortalization processes of cells as well as in human carcinogenesis.
MATERIALS AND METHODS
Cell Culture and Tissue Samples
The cell lines WI-38, RWPE-1, CCD-18Co, Hs 677.St, AGS, K-562, and SVG p12 were purchased from the American Type Culture Collection (Manassas, VA). WI-38 VA13, DU-145, KM1214, CCD-986sk, WM-266-4, NCI-H596, HeLa, SK-N-SH, A172, and WI-26 VA-4 were obtained from the Korean Cell Line Bank (Seoul, Korea). Senescent WI-38 cells were obtained by repeated passages until it reached senescence. WI-38, CCD-18Co, WM-266-4, and SVG p12 were propagated in minimal essential media containing Earle's salts, 2 mM L-glutamine, and 0.1 mM nonessential amino acids (EMEM), supplemented with 1 mM sodium pyruvate. WI-38 VA13, AGS, DU-145, NCI-H596, HeLa, SK-N-SH, and A172 were cultured in RPMI-1640; Hs 677.St, K-562, KM1214, CCD-986sk, and WI-26 VA-4 were maintained in Dulbecco's modified Eagle's Medium (DMEM); RWPE-1 was cultured in keratinocyte serum-free medium; all media were supplemented with 10% fetal bovine serum (FBS), streptomycin (100 µg/ml), and penicillin (100 U/ml). All cell lines were cultured at 37°C in a humidified 5.0% CO2 incubator. Tissue samples were obtained from Korea Lung Tissue Bank assigned and supported by the Korea Science & Engineering Foundation in the Ministry of Science & Technology. Tissue samples were homogenized in TRI REAGENT® (Molecular Research Center, Cincinnati, OH) and stored at −80°C.
Preparation of Total RNA
Total RNA was extracted from the cell lines and tissue samples with TRI REAGENT® according to the manufacturer's instructions. The A260 and A260/A280 of samples were determined with an ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., Waltham, MA). About 1 µg of total RNA was used for electrophoresis on a 1.0% agarose gel to verify its integrity.
Suppression Subtractive Hybridization (SSH)
SHH was performed with the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA) according to the protocol provided by the manufacturer. Pools of total RNA from senescent WI-38 and WI-38 VA13 were designated as driver and tester pools, respectively. The double-strand cDNAs were synthesized from 1 µg of total RNA and digested with RsaI to obtain shorter blunt-ended cDNA. The tester cDNA was subdivided into two populations and ligated to two different cDNA adaptors (adaptor 1 and 2R). Two hybridizations were performed with the tester and driver cDNAs; subsequently, the hybridized cDNAs were eliminated, leaving only the unhybridized (i.e., differentially expressed) cDNAs. The entire population of unhybridized molecules was then subjected to PCR to amplify the differentially expressed cDNAs. Only the molecules of the tester sample, which contained the two different adaptors, could be amplified exponentially. A second PCR amplification was performed using nested primers, in order to reduce background PCR products and to enrich the differentially expressed sequences.
Analysis of Subtraction Efficiency
Subtracted cDNA products were subjected to 18, 23, 28, or 33 cycles of PCR amplification with primers specific to glyceraldehyde 3-phosphate dehydrogenase (G3PDH), in order to evaluate subtraction efficiency. The sense (5′-ACC ACA GTC CAT GCC ATC AC-3′) and antisense (5′-TCC ACC ACC CTG TTG CTG TA-3′) G3PDH primers were provided in the PCR-Select cDNA Subtractive Kit. Unsubtracted controls were amplified in parallel to analyze the subtraction efficiency.
Cloning and Sequencing
The final subtraction PCR product was purified using a NucleoSpin Extraction kit (Clontech) and directly inserted into the pGEM-T Easy Vector System I (Promega Corp., Madison, WI). The ligation products were transformed into Escherichia coli strain DH5α. Plasmid DNA from each clone was purified using Wizard® Plus SV Miniprep DNA purification systems (Promega Corp.), and the presence of insert sequences was verified by digestion with EcoRI and sequencing using a primer specific to the T7 promoter and an ABI PRISM 3730XL Analyzer (Macrogen, Korea).
Bioinformatics Analysis
Sequences obtained from SSH were identified by comparing them to sequences in the National Center for Biotechnology Information (NCBI) database, using the Basic Local Alignment Search Tool (BLAST; www.ncbi.nlm.nih.gov/BLAST). Subcellular localization of the gene products were predicted by PSORT II (http://psort.ims.u-tokyo.ac.jp/form2.html).
Reverse Transcription-PCR Analysis
To validate differential expression among the cell lines and tissue samples, we performed reverse transcription polymerase chain reaction (RT-PCR). Full-length cDNA was synthesized using the SuperScript™ First-Strand Synthesis System (Invitrogen, Carlsbad, CA). All cDNAs used in this analysis were normalized with 18S rRNA, and primer pairs used for RT-PCR are listed in Table I and Supplementary Material 1. PCR was carried out in a total volume of 25 µl, using 1 µl of cDNAs, under the following conditions: 300 s at 94°C for initial denaturation, followed by 35 cycles of 94°C for 30 s (denaturation), 55°C for 30 s (annealing), and 72°C for 30 s (extension), with a final extension step at 72°C for 300 s. Aliquots (7 µl) of the PCR products were analyzed on 1% agarose gels. RT-PCR was performed at least three times.
| Genes | Primer sequences (5′–3′) | Size (bp) |
|---|---|---|
| ASPM | F: AGCCAACAAACAACAAGTTT | 409 |
| R: GGAGATTCGAGAAGAAAACA | ||
| RFC4 | F: GACCAAGGATCGAGGAGTA | 429 |
| R: GCAGCTGAGGTCATAGAATC | ||
| C3orf26 | F: GTTCTTGCAAAATCAGAACC | 602 |
| R: ACTTAGAAAAGGCCCAGTTT | ||
| BXDC2 | F: TTGGTCCTCCAAAACTTCCC | 179 |
| R: ACGGTATCATCCCAAAAGCC | ||
| C15orf44 | F: ACCGGCACCTCTGAGTAT | 512 |
| R: GTCTGAGTTGCTCATTCCAT | ||
| AURKA | F: TAGGAAGGTTATTGCACAGC | 402 |
| R: GCAAACAGTCTTAGGAATCG | ||
| C20orf77 | F: GAATATACTGCTGCCGCCAA | 299 |
| R: CACATCCTCCCAAAGTGTGC | ||
| RBMX | F: GAGATGTTTATTTGTCCCCA | 157 |
| R: TAGTCATCACGTGAACTGGA | ||
| CHA-V-97 | F: AGTTCAGGTTTTAAGGGAGG | 611 |
| R: CCTGGAGAAATTGACAGAAC | ||
| CHA-V-165 | F: ACCCAAAACAACAAGACATC | 687 |
| R: AGAATGTGTGTAGCTTGCCT | ||
| 18S rRNA | F: TACCTACCTGGTTGATCCTG | 243 |
| R: GGGTTGGTTTTGATCTGATA |
- F, forward direction; R, reverse direction.
Western Blotting
Protein extracts were prepared by PROPREP (Intron Inc., Korea) according to the manufacturer's instruction. After electrophoresis by 12% SDS–polyacrylamide gels, the separated proteins were blotted onto PVDF transfer membranes (Hybond-P; Amersham Biosciences). Blots were probed with rabbit polyclonal RFC4 IgG (1:2,000) and rabbit polyclonal AURKA IgG (1:2,000) for 1 h at room temperature, and mouse monoclonal BXDC2 (1:200) and goat polyclonal RBMX (hnRNP G) (1:1,000) for overnight at 4°C. Secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse IgG, or donkey anti-goat IgG. Rabbit anti-GAPDH was used for the normalization control. All antibodies used in this study were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Immunocomplexes were visualized with the Western Blotting Luminol Reagent (Santa Cruz Biotechnology) using the luminescent image analyzer LAS-3000 (FUJIFILM).
RESULTS
Efficiency of SSH
The efficiency of SSH was examined by PCR analysis with primers of G3PDH. The housekeeping gene displayed significant decrease in the amount of amplicon in subtracted cDNAs compared to unsubtracted cDNAs (Fig. 1A). Approximately seven to eight additional cycles of amplification were required for the subtracted cDNAs to meet the equivalent amount of G3PDH in the unsubtracted cDNAs (Fig. 1B). Subtracted library was successfully established.
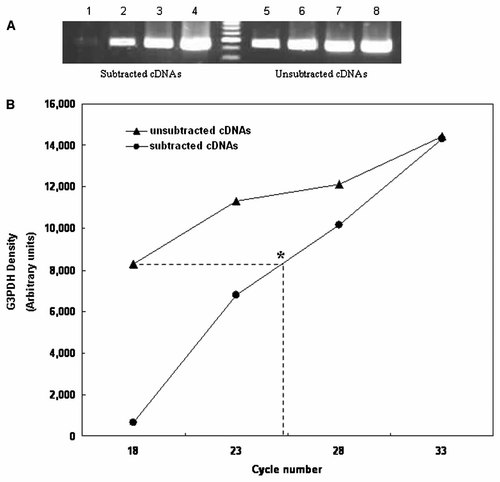
Subtraction efficiency of subtracted cDNAs. Subtraction efficiency was determined by analyzing the amount of remaining glyceraldehyde 3-phosphate dehydrogenase (G3PDH) with specific primers. A: PCR amplification was performed at 18, 23, 28, or 33 cycles. Lanes 1–4, subtracted cDNAs; lanes 5–8, unsubtracted cDNAs. Lanes 1 and 5 underwent 18 cycles; lanes 2 and 6, 23 cycles; lanes 3 and 7, 28 cycles; and lanes 4 and 8, 33 cycles. Lane M, marker (100 bp). B: The quantitative analysis of signal intensity for G3PDH. Additional cycles of amplification required for the subtracted cDNAs to obtain equivalent G3PDH signals in the 18 cycles of unsubtracted cDNAs is marked with asterisk (*).
Classification of Subtracted Genes
We cloned the SSH products and randomly selected 243 out of the resulting clones, and sequenced them. In 51 of 243 clones, the quality of the nucleotide sequence was not sufficient to yield significant data for this study; since these 51 clones were vector sequences, sequences with many ambiguities, and repetitive sequences, we excluded them from the analysis. We analyzed the remaining 192 out of 243 by comparing them to known NCBI Genbank sequences. As a result, 116 clones corresponded to 110 known genes and 6 uncharacterized known human genes. Among the remaining 76 clones, 33 clones corresponded only to ESTs, and 11 clones exhibited sequence similarity to genomic DNA sequences (Table II). Subsequent bioinformatics analysis revealed that the known genes were associated with diverse biological processes, such as cell cycle or proliferation, metabolism, cytoskeleton, transcription, cellular transport, signal transduction, and other functions; some corresponded to uncharacterized genes (Fig. 2, Table III).
| Category of cDNA | N | % |
|---|---|---|
| Total cDNAs generated | 243 | |
| cDNAs excluded from sequence database | 51 | |
| Subtracted cDNAs | 192 | 100 |
| Match with known genes | 110 | 57.3 |
| Match with uncharacterized genes | 6 | 3.1 |
| Match with SV40 | 14 | 7.3 |
| Only match with ETSs | 33 | 17.2 |
| Only match with human genomic sequences | 11 | 5.7 |
| Redundant results | 18 | 9.4 |
- N, number of clones.
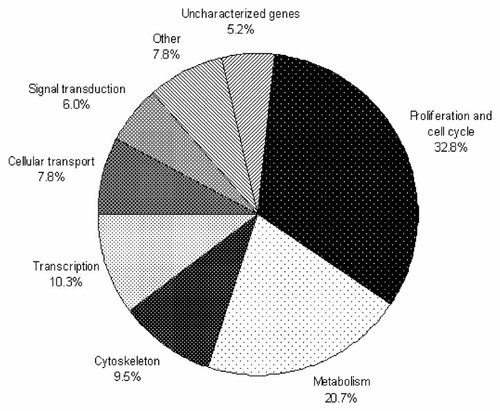
Functional classification of 116 differentially expressed known genes.
| GenBank ID | Gene symbol | Gene description | Ra | Predicted localizationb | Cytoband |
|---|---|---|---|---|---|
| Genes related to proliferation or cell cycle | |||||
| NM_005324 | H3F3B | H3 histone, family 3B (H3.3B) | 1 | M, N | 17q25 |
| NM_001067 | TOP2A | Topoisomerase (DNA) II alpha 170 kDa | 2 | N | 17q21–q22 |
| NM_181573 | RFC4 | Replication factor C (activator 1) 4 | 1 | C | 3q27 |
| NM_006283 | TACC1 | Transforming, acidic coiled-coil containing protein 1 | 1 | N | 8p11 |
| NM_013277 | RACGAP1 | Rac GTPase activating protein 1 | 1 | N | 12q13.13 |
| NM_198434 | AURKA | Aurora kinase A | 1 | N | 20q13.2-q13.3 |
| NM_031966 | CCNB1 | cyclin B1 | 1 | M | 5q12 |
| NM_003733 | OASL | 2'–5'-oligoadenylate synthetase-like | 1 | C | 12q24.2 |
| NM_006016 | CD164 | CD164 molecule, sialomucin | 1 | PM | 6q21 |
| NM_018685 | ANLN | Anillin, actin binding protein (scraps homolog, Drosophila) | 1 | N | 7p15-p14 |
| NM_005590 | MRE11A | Meiotic recombination 11 | 1 | N | 11q21 |
| NM_004612 | TGFBR1 | Transforming growth factor, beta receptor I (activin A receptor type II-like kinase, 53kDa) | 1 | ER | 9q22 |
| NM_152434 | CWF19L2 | CWF19-like 2 | 1 | N | 11q22.3 |
| NM_012325 | MAPRE1 | Microtubule-associated protein, RP/EB family, member 1 | 1 | C | 20q11.1-q11.23 |
| NM_018136 | ASPM | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) | 1 | N | 1q31 |
| NM_004396 | DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 1 | N | 17q21 |
| NM_004397 | DDX6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | 1 | C | 11q23.3 |
| NM_002687 | PNN | pinin, desmosome associated protein | 1 | N | 14q21.1 |
| NM_139323 | YWHAB | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 2 | N | 20q13.1 |
| AY796305.1c | NF1 | Neurofibromin 1 | 1 | M | 17q11.2 |
| NM_003384 | VRK1 | Vaccinia related kinase 1 | 1 | M | 14q32 |
| NM_021913 | AXL | AXL receptor tyrosine kinase | 1 | ER | 19q13.1 |
| NM_014720 | SLK | STE20-like kinase | 1 | N | 10q25.1 |
| NM_033306 | CASP4 | Caspase 4, apoptosis-related cysteine peptidase | 3 | N | 11q22.2-q22.3 |
| NM_003400 | XPO1 | Exportin 1 (CRM1 homolog, yeast) | 1 | C | 2p16 |
| NM_001316 | CSE1L | CSE1 chromosome segregation 1-like (yeast) | 1 | C | 20q13 |
| NM_006417 | IFI44 | Interferon-induced protein 44 | 1 | C | 1p31.1 |
| NM_003220 | TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | 2 | N | 6p24 |
| NM_005746 | NAMPT | Nicotinamide phosphoribosyltransferase | 1 | C | 7q22.2 |
| NM_001682 | ATP2B1 | ATPase, Ca++ transporting, plasma membrane 1 | 1 | PM | 12q21.3 |
| NM_017943 | FBXO34 | F-box protein 34 | 1 | N | 14q22.3 |
| NM_080927 | DCBLD2 | Discoidin, CUB and LCCL domain-containing 2 | 1 | PM | 3q12.1 |
| NM_002128 | HMGB1 | High-mobility group box 1 | 1 | N | 13q12 |
| NM_005504 | BCAT1 | Branched chain aminotransferase 1, cytosolic | 1 | C | 12pter-q12 |
| NM_003406 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 3 | N | 8q23.1 |
| NM_014781 | RB1CC1 | RB1-inducible coiled-coil 1 | 1 | N | 8q11 |
| NM_024408 | NOTCH2 | Notch homolog 2 (Drosophila) | 1 | EX, PM | 1p13-p11 |
| NM_006888 | CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | 1 | C | 14q24-q31 |
| Genes related to metabolism | |||||
| NM_005956 | MTHFD1 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 1 | C | 14q24 |
| NM_199229 | RPE | Ribulose-5-phosphate-3-epimerase | 1 | C | 2q32-q33.3 |
| NM_020186 | ACN9 | ACN9 homolog (S. cerevisiae) | 1 | M | 7q21.3 |
| NM_194318 | B3GALTL | Beta 1,3-galactosyltransferase-like | 1 | EX | 13q12.3 |
| NM_004458 | ACSL4 | acyl-CoA synthetase long-chain family member 4 | 1 | C | Xq22.3-q23 |
| NM_018638 | ETNK1 | Ethanolamine kinase 1 | 1 | M | 12p12.1 |
| NM_015423 | AASDHPPT | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | 1 | C | 11q22 |
| NM_005000 | NDUFA5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex | 1 | C | 7q32 |
| NM_005002 | NDUFA9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | 1 | C | 12p13.3 |
| NM_001685 | ATP5J | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 1 | M | 21q21.1 |
| NM_003680 | YARS | Tyrosyl-tRNA synthetase | 1 | C | 1p35.1 |
| NM_001634 | AMD1 | Adenosylmethionine decarboxylase 1 | 1 | C | 6q21-q22 |
| NM_006452 | PAICS | Phosphoribosylaminoimidazole carboxylase, phosphoribosy-laminoimidazole succinocarboxamide synthetase | 1 | C | 4q12 |
| NM_203464 | AK3L1 | Adenylate kinase 3-like 1 (AK3L1) | 3 | M | 1p31.3 |
| NM_000819 | GART | Phosphoribosylglycinamide formyltransferase, phosphorribosylglycinamide synthetase, phosphoribosylamino-imidazole synthetase | 1 | C | 21q22.11 |
| NM_014674 | EDEM1 | ER degradation enhancer, mannosidase alpha-like 1 | 1 | EX | 3p26.2 |
| NM_005530 | IDH3A | Isocitrate dehydrogenase 3 (NAD+) alpha | 1 | C | 15q25.1-q25.2 |
| NM_005028 | PIP4K2A | Phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | 1 | M | 10p12.2 |
| NM_003101 | SOAT1 | Sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 1 | PM | 1q25 |
| NM_004462 | FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 | 1 | C | 8p23.1-p22 |
| NM_017784 | OSBPL10 | Oxysterol binding protein-like 10 | 1 | N | 3p22.3 |
| NM_182796 | MAT2B | Methionine adenosyltransferase II, beta | 1 | C | 5q34-q35 |
| NM_002004 | FDPS | Farnesyl diphosphate synthase (farnesyl pyrophosphate syn-thetas dimethylallyltranstransferase, geranyltranstransferase) | 1 | M | 1q22 |
| NM_182729 | TXNRD1 | Thioredoxin reductase 1 | 1 | C | 12q23–q24.1 |
| Genes related to cytoskeleton | |||||
| NM_002906 | RDX | Radixin | 1 | N | 11q23 |
| NM_003628 | PKP4 | Plakophilin 4 | 2 | N | 2q23-q31 |
| NM_000381 | MID1 | Midline 1 (Opitz/BBB syndrome) | 1 | N | Xp22 |
| NM_003379 | EZR | Ezrin, transcript variant 2 | 1 | N | 6q25.2-q26 |
| NM_000210 | ITGA6 | Integrin, alpha 6 | 1 | G, PM, ER | 2q31.1 |
| NM_198086 | JUB | jub, ajuba homolog (Xenopus laevis) | 1 | C | 14q11.2 |
| NM_001547 | IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | 2 | N | 10q23-q25 |
| NM_003887 | DDEF2 | Development and differentiation enhancing factor | 1 | N | 2p25 |
| NM_014970 | KIFAP3 | Kinesin-associated protein 3 | 1 | C | 1q24.2 |
| NM_003816 | ADAM9 | ADAM metallopeptidase domain 9 (meltrin gamma) | 1 | ER | 8p11.23 |
| NM_002160 | TNC | Tenascin C (hexabrachion) | 1 | C | 9q33 |
| Genes related to transcription | |||||
| NM_002139 | RBMX | RNA binding motif protein, X-linked | 1 | N | Xq26.3 |
| NM_024721 | ZFHX4 | Zinc finger homeobox 4 | 1 | N | 8q21.11 |
| NM_004768 | SFRS11 | Splicing factor, arginine/serine-rich 11 (SFRS11) | 1 | N | 1p31 |
| NM_153207 | AEBP2 | AE binding protein 2 | 3 | N | 12p12.3 |
| NM_020235 | BBX | Bobby sox homolog | 1 | C | 3q13.1 |
| NM_001537 | HSBP1 | Heat shock factor binding protein 1 | 1 | N | 16q23.3 |
| NM_020948 | MIER1 | Mesoderm induction early response 1 homolog (Xenopus laevis) | 1 | EX | 1p31.3 |
| NM_015578 | LSM14A | LSM14A, SCD6 homolog A (S. cerevisiae) | 1 | N | 19q13.11 |
| NM_152344 | LSM12 | LSM12 homolog (S. cerevisiae) | 1 | N | 17q21.31 |
| NM_022491 | SUDS3 | Suppressor of defective silencing 3 homolog (S. cerevisiae) | 2 | N | 12q24.23 |
| NM_203350 | ZRANB2 | Zinc finger, RAN-binding domain containing 2 | 1 | N | 1p31 |
| NM_003137 | SRPK1 | SFRS protein kinase 1 | 1 | N | 6p21.3-p21.2 |
| Genes related to cellular transport | |||||
| NM_002264 | KPNA1 | Karyopherin alpha 1 (importin alpha 5) | 1 | C | 3q21 |
| NM_002266 | KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 1 | ER | 17q24.2 |
| NM_016079 | VPS24 | Vacuolar protein sorting 24 homolog (S. cerevisiae) | 1 | N | 2p24.3-p24.1 |
| NM_002271 | RANBP5 | RAN binding protein 5 | 1 | C | 13q32.2 |
| NM_006817 | ERP29 | Endoplasmic reticulum protein 29 | 1 | EX | 12q24.13 |
| NM_203437 | AFTPH | Aftiphilin | 1 | N | 2p14 |
| NM_006345 | SLC30A9 | Solute carrier family 30 (zinc transporter), member 9 | 1 | ER | 4p13-p12 |
| NM_020342 | SLC39A10 | Solute carrier family 39 (zinc transporter), member 10 | 1 | PM | 2q32.3 |
| NM_004896 | VPS26A | Vacuolar protein sorting 26 homolog A (yeast) | 1 | C | 10q21.1 |
| Genes related to signal transduction | |||||
| NM_015595 | SGEF | Src homology 3 domain-containing guanine nucleotide exchange factor | 1 | N | 3q25.2 |
| NM_005207 | CRKL | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | 1 | N | 22q11.21 |
| NM_014752 | SPCS2 | Signal peptidase complex subunit 2 homolog (S. cerevisiae) | 1 | ER | 11q13.4 |
| NM_005188 | CBL | Cas-Br-M (murine) ecotropic retroviral transforming sequence | 1 | N | 11q23.3 |
| NM_000902 | MME | Membrane metallo-endopeptidase | 1 | C, ER | 3q25.1-q25.2 |
| NM_002712 | PPP1R7 | Protein phosphatase 1, regulatory (inhibitor) subunit 7 | 1 | N | 2q37.3 |
| NM_003506 | FZD6 | Frizzled homolog 6 (Drosophila) | 1 | PM | 8q22.3-q23.1 |
| Genes related to other functions | |||||
| NM_001967 | EIF4A2 | Eukaryotic translation initiation factor 4A, isoform 2 | 1 | C | 3q28 |
| NM_018321 | BXDC2 | Brix domain containing 2 | 1 | N | 5p13.2 |
| NM_006392 | NOL5A | Nucleolar protein 5A (56kDa with KKE/D repeat) | 1 | N | 20p13 |
| EF194101d | HERV-H | Homo sapiens endogenous virus HERV-H | 1 | — | Xp22.32 |
| NM_002818 | PSME2 | Proteasome activator subunit 2(PA28 beta) | 1 | C | 14q11.2 |
| NM_006287 | TFPI | Tissue factor pathway inhibitor | 1 | EX | 2q32 |
| NM_001993 | F3 | Coagulation factor III (thromboplastin, tissue factor) | 1 | V, C | 1p22-p21 |
| NM_203331 | CD59 | CD59 molecule, complement regulatory protein | 1 | EX | 11p13 |
| NM_153810 | C10orf46 | Chromosome 10 open reading frame 46 | 1 | N | 10q26.11 |
| Uncharacterized genes | |||||
| NM_024056 | TMEM106C | Transmembrane protein 106C | 1 | ER, M | 12q13.1 |
| NM_014924 | KIAA0831 | Hypothetical protein LOC22863 | 1 | C | 14q22.3 |
| NM_021215 | C20orf77 | Chromosome 20 open reading frame 77 | 1 | N | 20q11.21-q12 |
| NM_032359 | C3orf26 | Chromosome 3 open reading frame 26 | 1 | N | 3q12.1 |
| NM_030800 | C15orf44 | Chromosome 15 open reading frame 44 | 1 | C | 15q22.31 |
| NM_030939 | C6orf62 | Chromosome 6 open reading frame 62 | 2 | C | 6p22.2 |
- a Recovery frequency.
- b Subcellular location was predicted using PSORT II [Horton and Nakai, 1997]. C, cytoplasm; EM, extracellular matrix; EX, extracellular; ER, endoplasmic reticulum; M, mitochondria; N, nucleus; PM, plasma membrane; G, Golgi; V, vesicles of secretory system.
- c The subtracted clone sequence was matched with NF1 gene but not with the mRNA sequence.
- d No CDS exists in HERV-H mRNA.
Expression of Known Genes in Immortalized Cells and Cancer Cells
We then examined differential expression of the selected genes (192 clones) between senescent and immortal lung cells using RT-PCR analysis. We chose 78 known genes and 2 novel genes that were strongly upregulated in the immortal lung cells (we additionally analyzed the expression levels of these 80 genes in WI-26 VA-4, NCI-H596, and SVG p12 cells) (Figs. 3A and 6A, Supplementary Material 2). We selected the genes located on chromosome 1q, 3q, 5p, 8q, 20q, and Xq that were previously reported as sites for frequent aberration in lung cancers [Pei et al., 2001; Wong et al., 2003]. Finally, we analyzed eight known genes and two novel genes by RT-PCR using diverse cell lines.
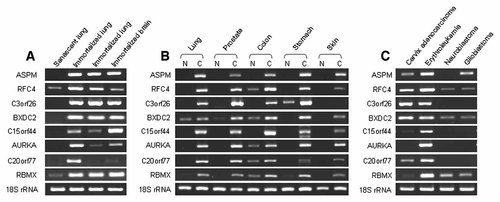
Differential mRNA expression of eight known genes in various cell lines by reverse transcription polymerase chain reaction (RT-PCR). A: Senescent and immortalized cell lines. Senescent lung (WI-38), SV40-immortalized lung (WI-38 VA13), SV40-immortalized lung (WI-26 VA-4), SV40-immortalized brain (SVG p12). B: Pairs of normal and cancer cell lines. Lung N (WI-38), C (adenosquamous, NCI-H596); prostate N (RWPE-1), C (adenocarcinoma, DU-145); colon N (CCD-18Co), C (carcinoma, KM1214); stomach N (Hs 677.St), C (adenocarcinoma, AGS); skin N (CCD-986sk), C (melanoma, WM-266-4). C: Cancer cell lines. Cervix adenocarcinoma (HeLa), erythroleukemia (chronic myelogenous leukemia, K562), neuroblastoma (SK-N-SH), glioblastoma (A172). N and C indicate normal and cancer, respectively.
As shown in Figure 3A, expression of 8 genes analyzed were strongly upregulated in immortalized cells. Among them, expression of C20orf77 was specifically upregulated only in immortalized lung cells (WI-38 VA13), but not in other cell lines that were immortalized by SV40. Expressions of eight known genes were dramatically increased in cancer cells from lungs, prostates, colons, and stomachs compared to those of normal cells (Fig. 3B). Skin cancer cells also highly expressed seven known genes, but not C3orf26 (Fig. 3B). Eight known genes showed intensive expression in erythroleukemia cells (Fig. 3C). All the known genes, except C15orf44, AURKA, and RBMX, were also upregulated in cervix adenocarcinoma cells (Fig. 3C). Only RFC4 and BXDC2 were expressed in both types of brain cancer (neuroblastoma and glioblastoma). Corresponding to previous report [Horvath et al., 2006], ASPM was overexpressed in glioblastoma cells but not in neuroblastoma (Fig. 3C).
Furthermore, we selected four representative genes to examine the expression level of proteins in diverse cell lines by Western blotting. As a result, RFC4, BXDC2, AURKA, and RBMX were mostly detectable in cancer cells and SV40-immortalized cells. We confirmed the protein levels upregulated in immortalized cells compared to senescent cells (Fig. 4A). Proteins were detectable in lung, prostate, and colon cancers but undetectable in stomach and skin cancers (Fig. 4B). These proteins were also detectable in other cancer cells, especially highly detectable in leukemia cells (Fig. 4C).
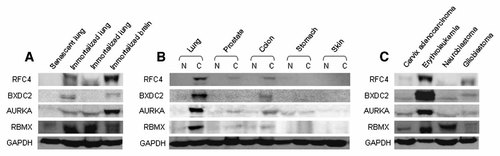
Differential protein expression of 8 known genes in various cell lines by Western blotting. A: Senescent and immortalized cell lines. Senescent lung (WI-38), SV40-immortalized lung (WI-38 VA13), SV40-immortalized lung (WI-26 VA-4), SV40-immortalized brain (SVG p12). B: Pairs of normal and cancer cell lines. Lung N (WI-38), C (adenosquamous, NCI-H596); prostate N (RWPE-1), C (adenocarcinoma, DU-145); colon N (CCD-18Co), C (carcinoma, KM1214); stomach N (Hs 677.St), C (adenocarcinoma, AGS); skin N (CCD-986sk), C (melanoma, WM-266-4). C: Cancer cell lines. Cervix adenocarcinoma (HeLa), erythroleukemia (chronic myelogenous leukemia, K562), neuroblastoma (SK-N-SH), glioblastoma (A172). N and C indicate normal and cancer, respectively.
Expressions of these genes were further analyzed in two types of non-small-cell lung carcinoma tissues, squamous cell carcinoma and adenocarcinoma. Not all eight genes were upregulated in both types of cancer tissues compared to corresponding normal tissues. However, the mRNA expression level of BXDC2 and RBMX were strongly upregulated in most of the samples (Fig. 5).

Validation of known gene mRNA expression levels by RT-PCR in normal lung and non-small-cell lung cancers (NSCLCs) tissues. A: Pairs of normal lung and squamous cell carcinoma tissues. B: Pairs of normal and adenocarcinoma tissues. N and T indicate normal and tumor tissues, respectively.
Identification of 2 Novel Genes in the Subtracted cDNA Library
Among the unknown genes, we identified two clones intensively upregulated in immortalized cells. We named the ESTs (expressed sequence tags) as CHA-V-97 and CHA-V165, respectively. CHA-V-97 (Genbank accession number: GH270327) locates on Xp22 and CHA-V-165 (Genbank accession number: GH270328) locates on chromosome 12. The mRNA expression patterns of the two novel genes were similar to those of known genes. Immortalized cells expressed CHA-V-97 (except in SVG p12) and CHA-V-165 more strongly than senescent cells (Fig. 6A). Similarly, expression of CHA-V-165 was highly upregulated in cancer cells than those of normal cells but CHA-V-97 showed upregulation in lungs and prostates pairs (Fig. 6B). Upregulation of these clones were also detected in other cancer cell lines (Fig. 6C). Furthermore, expression of both clones was also seemingly upregulated in tissues from both types of lung cancers, SCC & AC, compared to those of normal lung tissues (Fig. 7).
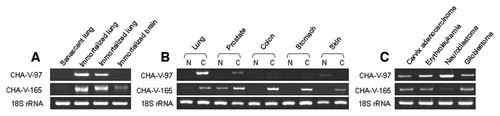
Differential mRNA expression of two novel genes in various cell lines by reverse transcription polymerase chain reaction (RT-PCR). A: Senescent and immortalized cell lines. Senescent lung (WI-38), SV40-immortalized lung (WI-38 VA13), SV40-immortalized lung (WI-26 VA-4), SV40-immortalized brain (SVG p12). B: Pairs of normal and cancer cell lines. Lung N (WI-38), C (adenosquamous, NCI-H596); prostate N (RWPE-1), C (adenocarcinoma, DU-145); colon N (CCD-18Co), C (carcinoma, KM1214); stomach N (Hs 677.St), C (adenocarcinoma, AGS); skin N (CCD-986sk), C (melanoma, WM-266-4). C: Cancer cell lines. Cervix adenocarcinoma (HeLa), erythroleukemia (chronic myelogenous leukemia, K562), neuroblastoma (SK-N-SH), glioblastoma (A172). N and C indicate normal and cancer, respectively.
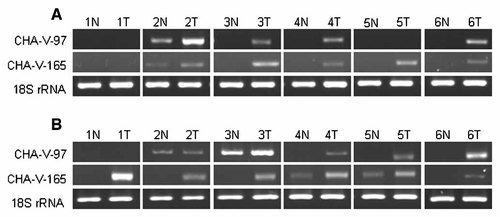
Validation of two novel gene mRNA expression levels by RT-PCR in normal lung and non-small-cell lung cancers (NSCLCs) tissues. A: Pairs of normal lung and squamous cell carcinoma tissues. B: Pairs of normal and adenocarcinoma tissues. N and T indicate normal and tumor tissues, respectively.
DISCUSSION
Several methods have been used to examine differentially expressed genes, including differential display polymerase chain reaction (DD-PCR) [Liang and Pardee, 1992], RNA fingerprinting with random-primed PCR [Perucho et al., 1995], representational difference analysis (RDA) [Lisitsyn and Wigler, 1993], expressed sequenced tag (EST) database analysis [Vasmatzis et al., 1998], DNA microarray [Schena et al., 1995], and suppression subtractive hybridization (SSH) [Diatchenko et al., 1996]. Among these several methods, SSH has an advantage to identify the genes that are definitely unknown.
In order to identify differentially expressed novel genes in immortalized cells, we performed SSH on cDNA set, SV40-immortalized lung and senescent lung fibroblasts. As a result, we obtained 79.0% (192 out of 243) positive clones with only 9.4% redundancy (15 redundant clones in known genes and 3 in unknown genes). We analyzed the sequence similarity to the NCBI Genbank sequences. As a result, we discovered 2 novel genes, both of which were highly upregulated in the immortal cells.
Moreover, RT-PCR analysis revealed that most of the known genes exhibited expression differences between senescent and immortalized cells, in which expression of 78 clones (66.7%) were highly distinguishable. Because immortalization process is carried out with indefinite proliferation, it was anticipated that the genes with different expression may be involved in cell cycle, growth, and division. Consistent with this prediction, 38 of the known cDNAs (32.8% of total known genes) identified by SSH were related to cell cycle or proliferation, and 81.6% (31 out of 38 known genes) among these genes were strongly upregulated in the immortalized cells. Taken as a whole, these results demonstrate that the screening process subtracted immortalization-associated genes as expected.
According to the assumption that immortalization is an early event for carcinogenesis, we assumed that the majority of these genes may be associated with cancer development as well as immortalization. Results from previous studies have shown that aberrant chromosome gains at 1q, 3q, 5p, 8q, 12q, 14q, 17q, 20q, and Xq were frequently detectable in lung cancers, especially in NSCLC [Pei et al., 2001; Wong et al., 2003]. We selected several genes located on the sites: ASPM (1q31), RFC4 (3q27), C3orf26 (3q12.1), BXDC2 (5p13.2), C15orf44 (15q22.31), AURKA (20q13.2-q13.3), C20orf77 (20q11.21-q12), and RBMX (Xq26.3).
There are evidences that these known genes might be associated with human cancers. ASPM, abnormal spindle-like microcephaly associated, is essential for formation of proper mitotic spindle in mitosis and meiosis [Saunders et al., 1997]. Inhibition of ASPM mediated by siRNA-knockdown in glioblastoma inhibits proliferation of tumor and neural stem cell [Horvath et al., 2006]. RFC4, replication factor C, is responsible for the checkpoint pathways in DNA damage [Ellison and Stillman, 2003]. Previous reports have shown that expression of RFC4 was upregulated in head and neck squamous cell carcinoma (HNSCC), cervical cancers, and breast cancers [Slebos et al., 2006; Wen et al., 2006; Narayan et al., 2007]. BXDC2, brix domain containing 2, is associated with ribosome biogenesis. However, studies on BXDC2 were rarely done in human cancers. The upregulated expression of BXDC2 in human cancers in this study indicates that activation of ribosome biogenesis is necessary for formation and/or survival of tumors. Aurora kinase A (AURKA) is a cell cycle-regulated kinase contributing to the formation of microtubules. Overexpression of AURKA in diploid human breast epithelial cells induced centrosome abnormality, leading to aneuploidy in various types of human cancers [Zhou et al., 1998]. AURKA is frequently amplified and overexpressed in human cancers [Sen et al., 1997; Sakakura et al., 2001; Gritsko et al., 2003; Li et al., 2003]. AURKA also plays an important role on ovarian carcinogenesis [Chung et al., 2005]. Recent studies of RBMX, RNA binding motif X-linked, reveal that RBM proteins may play a role in apoptosis modulation and in angiogenesis in aspect that RBMX correlates with CD105, a gene involved in oncogenic development of breast cancers [Sutherland et al., 2005].
There were three genes, C3orf26, C15orf44, and C20orf77, of which biological functions remain unclear. Their elevated expression in SV40-immortalized cells, cancer cells, and NSCLC tissues may also suggest their potential associations with immortalization processes.
Based on the reports of gene association study in human cancers, we analyzed their expression in diverse cell lines and lung tissues of non-small-cell lung cancer patients. Their mRNA expressions were highly upregulated in various cancer cells and SV40-immortalized cells. The protein levels of the four representative genes (RFC4, BXDC2, AURKA, and RBMX) were strongly detectable in cancer or immortalized cells compared to the normal cells. However, we observed that the protein expression patterns have slight difference to their mRNA expression levels. These proteins were highly detectable in lung cancer cells and leukemia cells. However, expressions were almost undetectable in stomach and skin cancer. These results indicate that such proteins specifically function according to cancer types. Another possibility is that there might be translational regulation of these four genes in those cancer types. The mRNA levels of the eight genes we examined also have upregulated pattern of mRNA levels in the tissues of non-small-cell lung cancers compared to the normal type tissues. These results demonstrate that these candidate genes play a role in lung cancers.
The two novel genes, CHA-V-97 and CHA-V-165, were also upregulated in SV40-immortalized cells, cancer cells, and NSCLCs tissues. The full transcript remains to be identified. Further studies of these genes might suggest clues of the unknown mechanisms underlying the immortalization processes and carcinogenesis in human cancers.
In summary, our SSH analysis identified a population of genes from immortalized cells which included many known genes, several uncharacterized genes and novel genes. We selected genes that were located on the overrepresented chromosome (1q, 3q, 5p, 8q, 20q, and Xq) in NSCLCs. Among them, ASPM (1q), RFC4 (3q), C3orf26 (3q), BXDC2 (5p), AURKA (20q), C20orf77 (20q), and RBMX (Xq) were highly upregulated in SV40-immortalized cells, cancer cells, and NSCLC tissues. Furthermore, two novel genes, CHA-V-97 and CHA-V-165, were also upregulated in various cancers as well as immortalized cells in human. Taken together, we confirmed upregulation of genes specific for immortalized cells and cancer cells, suggesting that they may play a role on immortalization or carcinogenesis in human cancers.
Acknowledgements
This work was supported by a grant from the Stem Cell Research Program (2006-04127) of the Ministry of Education, Science, and Technology, and Korea Science and Engineering Foundation.




