Isoform-specific cleavage of 14-3-3 proteins in apoptotic JURL-MK1 cells
Abstract
The proteins of 14-3-3 family are substantially involved in the regulation of many biological processes including the apoptosis. We studied the changes in the expression of five 14-3-3 isoforms (β, γ, ε, τ, and ζ) during the apoptosis of JURL-MK1 and K562 cells. The expression level of all these proteins markedly decreased in relation with the apoptosis progression and all isoforms underwent truncation, which probably corresponds to the removal of several C-terminal amino acids. The observed 14-3-3 modifications were partially blocked by caspase-3 inhibition. In addition to caspases, a non-caspase protease is likely to contribute to 14-3-3's cleavage in an isoform-specific manner. While 14-3-3 γ seems to be cleaved mainly by caspase-3, the alternative mechanism is essentially involved in the case of 14-3-3 τ, and a combined effect was observed for the isoforms ε, β, and ζ. We suggest that the processing of 14-3-3 proteins could form an integral part of the programmed cell death or at least of some apoptotic pathways. J. Cell. Biochem. 106: 673–681, 2009. © 2009 Wiley-Liss, Inc.
The proteins of 14-3-3 family are ubiquitously expressed in all eukaryotic cells and involved in the regulation and coordination of many important biological processes [Masters and Fu, 2001; Mackintosh, 2004; Pozuelo Rubio et al., 2004; Aitken, 2006; Gardino et al., 2006; Porter et al., 2006]: among others, they prevent undesirable triggering of apoptotic cell death programs and help to maintain the correct progression of the cell cycle. In humans, the family consists of seven distinct isoforms (β/α, γ, ζ/δ, ε, η, σ, and τ/θ) with tissue-specific expression. The molecular weight (MW) of these acidic proteins is 27–28 kDa, with the exception of the isoform ε, which is somewhat larger (29 kDa). They form homo- or heterodimers which can bind one or two client proteins, mostly through a phosphoserine/phosphothreonine containing motif, although non-phosphorylated binding sites were also identified (e.g., on the proapoptotic protein Bax). The binding site is usually internal, but it can also be located at the C-terminus of the client protein [Coblitz et al., 2006]. Each 14-3-3 monomer comprises nine antiparallel α-helices and two subunits associate through their N-terminal domains to form a flattened U-shaped structure with two amphipathic grooves for ligand binding. While the inner surface of the dimer is composed of highly conserved residues, the C-termini of the monomers are the most divergent regions in sequence and adopt no strictly ordered structure. The affinity of 14-3-3 proteins for their targets is regulated by phosphorylation, both on the target proteins and on 14-3-3's themselves.
Although the high-affinity binding motifs are recognized by all 14-3-3 proteins, the different mammalian 14-3-3 isotypes appear to have specific functions [Kjarland et al., 2006; Jagemann et al., 2008; Niemantsverdriet et al., 2008]. However, despite the large number of known 14-3-3 targets, little data are available as to the specificity in substrate recognition and/or independent regulation of the expression level and activity for the individual 14-3-3 isoforms.
Binding of client proteins to 14-3-3s can have multiple effects including changes in enzymatic activity, subcellular localization, and protein complex formation. Figure 1 shows some of the well-documented interaction points between 14-3-3 proteins and the apoptotic machinery [Bonnefoy-Berard et al., 1995; Bae et al., 2001; Brunet et al., 2002; Nomura et al., 2003; Won et al., 2003; Zhang et al., 2003, 2007; Tsuruta et al., 2004; Pendergast, 2005; Yoshida et al., 2005; Malissein et al., 2006; Qi et al., 2006; Lee et al., 2008; Yan et al., 2008]. Signals arriving from growth factor receptors activate kinases (Akt, PKA, etc.) which phosphorylate number of proapoptotic actors (e.g., Bad, FKHRL1, FOXO1, etc.) and thereby promote their binding to 14-3-3 proteins and inactivation. On the other hand, pro-death signals activate among others the kinase JNK which phosphorylates some of 14-3-3 isoforms and induces the release of proapoptotic client proteins (Bax, c-Abl, etc.) which can thus trigger the apoptosis. In a similar way, the family of 14-3-3 proteins interacts with multiple proteins which are essentially involved in the regulation of the cell-cycle progression, cell adhesion and migration, stress response, and other fundamental processes [Jin et al., 2004; Hermeking and Benzinger, 2006].
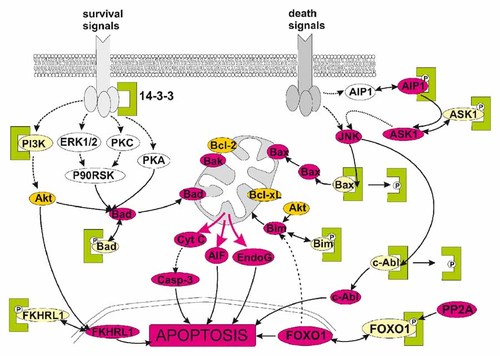
Some of the known interaction points between 14-3-3 protein family (green clamps) and the proteins operative during the apoptosis. Pro-apoptotic forms of the proteins are shown in violet, while the inactive forms are in light yellow.
In this work, we describe changes in the expression level of five 14-3-3 isoforms induced by the treatment of human leukemic cells with the kinase inhibitor Imatinib mesylate. JURL-MK1 [Di Noto et al., 1997] and K562 cell lines express the aberrant protein Bcr-Abl which is the primary cause of chronic myelogenous leukemia (CML) and whose inhibition by Imatinib mesylate results in apoptosis induction. We show that the lowered expression and cleavage of 14-3-3 proteins occurring during the apoptosis are to a large extent independent of caspase-3 activity and probably involve a non-caspase protease. The relative contributions of caspase-3-dependent and -independent mechanisms vary among the individual 14-3-3 isoforms.
MATERIALS AND METHODS
Chemicals
Imatinib mesylate was kindly provided by Novartis (Basel, Switzerland), 17- (allylamino)-17-desmethoxygeldanamycin (17-AAG) was purchased from Alexis (Lausen, Switzerland). The irreversible caspase inhibitors zDEVD-fmk and zVAD-fmk were from Sigma–Aldrich (Prague, Czech Republic) and Alexis, respectively. The antibodies against 14-3-3 β (sc-1657 recognizing N-terminus and sc-25276 recognizing C-terminus) and ε (sc-1020) were obtained from Santa Cruz Biotechnology (CA), anti-14-3-3 τ (T5942) was from Sigma–Aldrich, anti-14-3-3 ζ (AF2669) was purchased from R&D Systems (Germany), anti-14-3-3 γ (05-639) from Millipore (Upstate), the antibodies against 14-3-3 ε (MAB3053) and C-terminus of 14-3-3 ε (AB9732) from Millipore (Chemicon). Anti-caspase-3 antibody (sc-7272) recognizing both procaspase-3 and the active caspase-3 was obtained from Santa Cruz Biotechnology.
Cell Culture
JURL-MK1 cell line was obtained from DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany), and K562 cell line was from the European Collection of Animal Cell Cultures (Salisbury, UK). The cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2 humidified atmosphere. They were diluted to a density of (2–4) × 105 cells/ml three times a week.
2D Electrophoresis
The method for protein separation by 2D electrophoresis was described in detail previously [Grebeňová et al., 2006]. In brief, control and treated cells were lysed in Triton X-100 containing buffer and the lysates were subjected to isoelectric focusing followed by SDS–PAGE electrophoresis. The gels were stained with Coomassie blue, scanned, and analyzed employing Phoretix® 2D Expression Software (Nonlinear Dynamics, UK). The protein spots, which were repeatedly found to be altered by the treatment, were excised from the gel and subjected to MALDI–TOF mass spectrometry (MS) analysis.
Enzymatic In-Gel Digestion
Coomassie blue-stained protein spots were excised from the gel, cut into small pieces, and washed with 0.1 M 4-ethylmorpholine acetate (pH 8.1) in 50% acetonitrile (MeCN). After complete destaining, the gel was washed with water, shrunk by dehydration in MeCN, and reswelled again in water. The supernatant was removed and the gel was partly dried in a SpeedVac concentrator. The gel pieces were then reconstituted in a cleavage buffer containing 25 mM 4-ethylmorpholine acetate, 5% MeCN, and sequencing grade trypsin (20 ng/µl; Promega) or Asp-N protease (10 ng/µl; Roche Diagnostics GmbH, Mannheim, Germany). After overnight digestion, the resulting peptides were extracted to 40% MeCN/0.1% TFA.
MALDI–TOF Mass Spectrometry (MS) and Protein Identification
An aqueous 50% MeCN/0.1% TFA solution of α-cyano-4-hydroxycinnamic acid (Sigma; 5 mg/ml) or 2,5-dihydroxybenzoic acid (Sigma; 20 mg/ml) was used as a MALDI matrix. The sample (0.5 µl) was deposited on the MALDI target and allowed to air-dry at room temperature. After complete evaporation, 0.5 µl of the matrix solution was added. MALDI mass spectra were measured on an Ultraflex III instrument (Bruker Daltonics, Bremen, Germany) equipped with a Smartbeam™ solid-state laser and LIFT™ technology for MS/MS analysis. The spectra were acquired in the mass range of 700–5,000 Da and calibrated externally using peptide calibration mix II (Bruker Daltonics).
The peak lists created using flexAnalysis 3.0 program (Bruker Daltonics) were subjected to database search employing in-house MASCOT program (Matrixscience). The following search settings were used: human subset of SwissProt 55.5 database; enzyme: trypsin or Asp-N ambic; variable modifications: acetyl (Protein N-term), carbamidomethyl (C), oxidation (M); MS mass tolerance: 50 ppm; max. missed cleavage: two for trypsin and five for Asp-N.
1D Electrophoresis and Western Blotting
The SDS electrophoresis and Western blotting were performed using the protocols which were described previously [Kuželová et al., 2007]. The protein concentration was measured using a BioRad protein assay (Bio-Rad, USA). As a rule, 10 µg of total protein was applied to each well. Actin band was used as a control of equal protein loading.
Caspase Activity Assessment
The activity of caspases was determined by fluorometric measurement of the kinetics of 7-amino-4-trifluoromethyl coumarin (AFC) release from the fluorogenic substrates Ac-DEVD-AFC, Ac-LEHD-AFC (both Sigma–Aldrich) and Ac-VDVAD-AFC (Alexis Biochemicals, CA), respectively, in the presence of the cell lysates. The method was described in detail in Kuželová et al. 2007. When used, the caspase inhibitors zDEVD-fmk or zVAD-fmk at the indicated concentration (0–50 µM) were added to the cells in culture simultaneously with the apoptosis inducer. After the incubation time, the cells were washed and lysed, the aliquots of cytosolic fraction were mixed with the appropriate fluorogenic substrate, and the linear increase of fluorescence intensity at 520 nm was monitored using Fluostar Galaxy microplate reader (BMG Labtechnologies, Germany).
Apoptosis Quantification
The fraction of cells containing apoptotic DNA breaks was measured by TUNEL assay using the In Situ Cell Death Detection Kit, Fluorescein (Roche Diagnostics GmbH) following the standard manufacturer's protocol. The extent of DNA labeling with fluorescein-dUTP was determined employing the Coulter Epics XL flow cytometer.
RESULTS
We have previously shown that Imatinib mesylate treatment induces apoptosis in JURL-MK1 cells [Kuželová et al., 2005]. Figure 2 shows the kinetics of Imatinib-induced caspase-3 activation measured using the fluorogenic caspase-3 substrate (panel A) and apoptotic DNA fragmentation detected by TUNEL method (panel B). We subsequently performed proteomic analysis of Imatinib effects on JURL-MK1 cell line by means of 2D electrophoresis and MALDI–TOF MS. Among multiple proteins which were altered by Imatinib treatment, some spots were identified as members of 14-3-3 protein family (Fig. 3A). The treatment resulted in a marked decrease of the spot intensities corresponding to 14-3-3 ε, τ, β + ζ (overlapping spots of β and ζ isoforms) and γ + ζ. Additionally, as a result of Imatinib treatment new spots identified as 14-3-3 ε, τ, β, and γ appeared at lower MWs suggesting possible fragmentation of the intact 14-3-3 proteins. Figure 3B shows the comparison of sequence coverage obtained using MALDI peptide mass mapping of two spots which were identified as 14-3-3 τ. The MALDI data of the spot corresponding to the intact form of 14-3-3 τ cover both N- and C-terminus of the protein while the C-terminal peptide was missing from the presumed fragment.
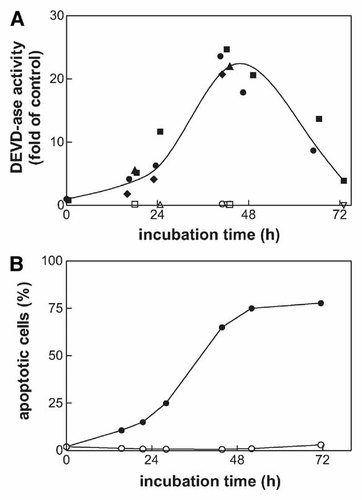
Caspase-3 activation and DNA fragmentation in JURL-MK1 cells treated with Imatinib mesylate. The cells were incubated with 1 µM Imatinib for up to 72 h. The activity of caspase-3 was measured using the fluorogenic substrate Ac-DEVD-AFC and expressed in relative units (ratio of values obtained from the treated samples and from the control). The results shown in panel A are merged from several independent experiments (different closed symbols). In some experiments, the caspase inhibitor zDEVD-fmk at 5 µM concentration was added to cells simultaneously with Imatinib (open symbols in panel A). The fraction of cells with apoptotic DNA was quantified by means of TUNEL method (panel B, open symbols represent untreated control). Equivalent results were obtained from repeated experiments.
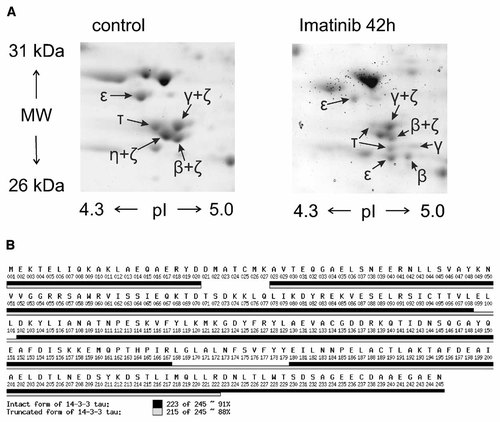
A: Sections of 2D electrophoretic maps showing the spots which were assigned to 14-3-3 proteins by mass spectrometry. JURL-MK1 cells were untreated (left panel) or treated with 1 µM Imatinib mesylate for 42 h (right panel). The spots labeled as β + ζ, γ + ζ, and η + ζ were identified as a mixture of these 14-3-3 isoforms. B: Comparison of MALDI peptide mass mapping data of two spots identified as 14-3-3 τ. The sequence coverage for both forms of the protein was calculated from both tryptic and Asp-N peptide mass maps. Black bars: intact 14-3-3 τ; clear bars: lower MW form of 14-3-3 τ.
Changes in the expression levels of 14-3-3 proteins were further studied by immunoblotting using specific antibodies against the isoforms ε, β/α, γ, τ, and ζ/δ. The Western blots contained no unspecific bands and the attribution of 28–29 kDa bands to 14-3-3 isoforms could be made unambiguously. As it is shown in Figure 4, the amount of all these 14-3-3 isoforms was lowered during the apoptosis induced by Imatinib mesylate. In parallel, we detected a progressive appearance of fragments of 14-3-3 β, γ, ε, and τ at about 27 kDa. These fragments were not detected when we used antibodies specific for C-terminal sequences of 14-3-3 ε and β (Fig. 4, right part). In the case of ζ isoform, the amount of fragment formed as a result of Imatinib treatment was low compared to the full-length protein and in some experiments was not detected at all.
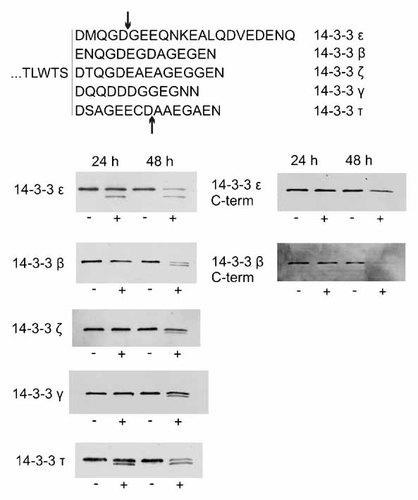
Top: alignment of C-terminal sequences of all studied 14-3-3 isoforms. The arrows point to the previously reported cleavage sites for caspase-3 on 14-3-3 ε and τ. Bottom: processing of 14-3-3 protein isoforms in JURL-MK1 cells treated with Imatinib mesylate. The cells were treated by 1 µM Imatinib for 24 or 48 h as indicated. Cell lysates from control cells (−) and from treated samples (+) were subjected to 1D electrophoresis and the expression pattern of the specified 14-3-3 isoforms was explored by means of immunoblotting using the corresponding antibody.
As some 14-3-3 isoforms had been shown to be substrates for caspase-3 [Nomura et al., 2003; Won et al., 2003], we analyzed the role of caspases in the observed 14-3-3 processing using irreversible caspase inhibitors: zDEVD-fmk as a relatively specific inhibitor of caspase-3 and the general caspase inhibitor zVAD-fmk. These compounds are able to penetrate the cell membrane and they bind covalently to the active site of caspases providing thereby the irreversible inhibition of caspase activity. We have previously shown by fluorometric monitoring of caspase-3 activity and by anti-caspase-3 immunoblotting that both these inhibitors are fully efficient at concentrations lower than 5 µM [Kuželová et al., 2007]. We verified that in the presence of 5 µM zDEVD-fmk or 5 µM zVAD-fmk, the DEVDase activity was maintained well under the control value (untreated JURL-MK1 cells) for up to 72 h following sample treatment with Imatinib mesylate (Fig. 2 A, open symbols). We also tested the effects of these inhibitors on the rate of cleavage of the fluorogenic peptide substrates Ac-LEHD-AFC and Ac-VDVAD-AFC, which are efficiently cleaved by caspase-9 and -2, respectively. As shown in Figure 5, Imatinib mesylate induced about 20-, 2-, and 4-fold increase in the cleavage rate of Ac-DEVD-AFC, Ac-LEHD-AFC, and Ac-VDVAD-AFC, respectively, and this increase was completely inhibited by simultaneous addition of 0.5 µM zDEVD-fmk. On the other hand, the inhibition of caspases under these conditions had only partial effect on the changes in 14-3-3 protein levels induced by Imatinib mesylate (Fig. 6A, lanes 3 and 5). The caspase inhibitors fully prevented 14-3-3 processing only in the case of γ isoform. The isoform 14-3-3 ε was responsive to caspase-3 inhibition, but we observed a large variability in the extent of 14-3-3 ε cleavage inhibition by zDEVD-fmk in individual experiments. The intensity of the fragment band was reduced to 3–77% of the original value (mean reduction to 39% from eight experiments) using 5 µM zDEVD-fmk. In the case of 14-3-3 β, the cleavage was reduced by zDEVD-fmk but significant amount of 14-3-3 fragment was still present in all experiments even at 50 µM inhibitor. Finally, the cleavage of 14-3-3 τ was almost unaffected by zDEVD-fmk in repeated experiments, even at 50 µM concentration (Fig. 6A, lane 4). The broad-range caspase inhibitor zVAD-fmk at 50 µM (but not 5 µM) concentration was able to prevent the changes in the expression level of all the studied 14-3-3 isoforms (Fig. 6A, lane 6) except for 14-3-3 τ.
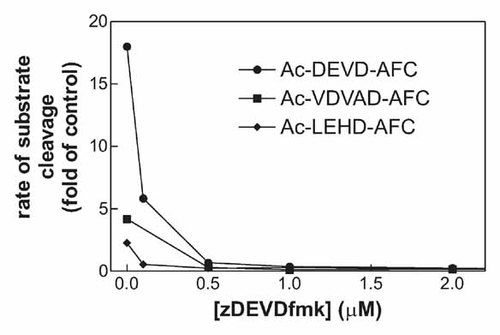
Effect of the caspase inhibitor zDEVD-fmk on the cleavage rate of fluorogenic caspase substrates Ac-DEVD-AFC, Ac-LEHD-AFC, and Ac-VDVAD-AFC. JURL-MK1 cells were treated with 1 µM Imatinib mesylate in combination with zDEVD-fmk at the indicated concentration for 43 h. Thereafter, they were washed in PBS, lysed, and an aliquot of the cytosolic extract was assayed for the cleavage activity. The results are expressed in relative units (ratio of activity values obtained from the treated samples and from the corresponding controls).
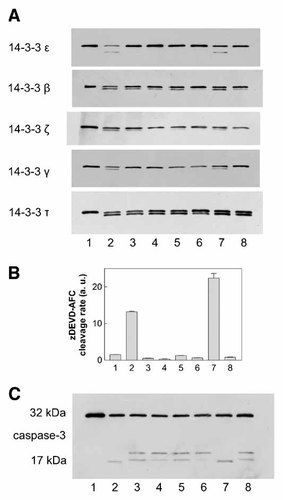
Effect of irreversible caspase inhibitors on 14-3-3 processing in JURL-MK1 cells treated with Imatinib mesylate or 17-AAG. The cells were treated with 1 µM apoptosis inducer alone or in combination with zDEVD-fmk or zVAD-fmk for 48 h. Sample aliquots were lysed and subjected to 1D electrophoresis and the expression pattern of the specified 14-3-3 isoforms was explored by means of immunoblotting using the corresponding antibody (panel A). Lane 1: controls; lane 2: Imatinib; lanes 3 and 4: Imatinib with zDEVD-fmk 5 µM or 50 µM, respectively; lanes 5 and 6: Imatinib with zVAD-fmk 5 µM or 50 µM, respectively; lane 7: 17-AAG; lane 8: 17-AAG with 5 µM zDEVD-fmk. In parallel, the samples were assessed for caspase-3 activity using the fluorogenic caspase-3 substrate zDEVD-AFC (panel B) and for procaspase-3 processing using anti-caspase-3 antibody (panel C). All results presented in this figure were obtained from the same set of experimental samples. Additional independent experiments yielded similar results.
We also examined the effects of an alternative apoptosis inducer, 17-AAG, an inhibitor of the heat shock protein HSP90, on the expression level of 14-3-3 proteins. The treatment of JURL-MK1 cells with 1 µM 17-AAG resulted in caspase-3 activation closely similar to that induced by Imatinib mesylate (data not shown) as well as in 14-3-3 cleavage (Fig. 6, lane 7). The MW of these fragments was apparently the same as in the case of Imatinib mesylate. The effect of caspase inhibition by 5 µM zDEVD-fmk during 17-AAG treatment was also isoform-dependent: while the inhibitor prevented or reduced the cleavage of 14-3-3 γ, ε, β, and ζ, it was not able to inhibit the cleavage of 14-3-3 τ (Fig. 6, lane 8).
The panels B and C of Figure 6 document caspase-3 inhibition by zDEVD-fmk and zVAD-fmk in the samples analyzed in the panel A of the figure. They show the rate of in vitro cleavage of the fluorogenic caspase-3 substrate and anti-caspase-3 immunoblots, respectively.
Despite the complete inhibition of caspase-3 activity, the effect of caspase inhibitors on the apoptosis progression was also very limited. The apoptotic DNA fragmentation induced by Imatinib was considerably reduced only in the presence of 50 µM zVAD-fmk (Fig. 7).
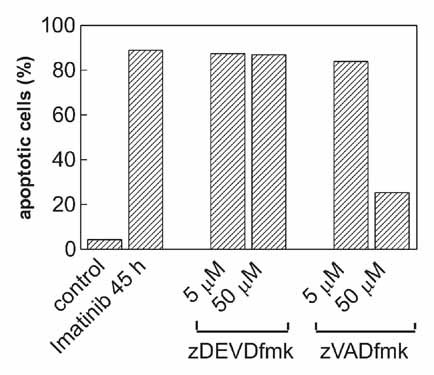
Effect of irreversible caspase inhibitors on the extent of apoptosis in JURL-MK1 cells treated with Imatinib mesylate. The cells were treated with 1 µM Imatinib mesylate alone or in combination with zDEVD-fmk or zVAD-fmk for 45 h as indicated and the fraction of cells with apoptotic DNA breaks was measured using TUNEL method. Two additional experiments yielded closely similar results.
We performed similar analysis of the effects of caspase inhibitors on 14-3-3 expression and on the apoptosis progression using the cell line K562 which is also derived from a patient with CML. Imatinib mesylate treatment of K562 cells led to similar changes in anti-14-3-3 immunoblots as in the case of JURL-MK1 cells (i.e., lowered expression and fragmentation of individual 14-3-3 isoforms). While the inhibitor zDEVD-fmk appeared to be to some extent more efficient in K562 cells (compared to JURL-MK1) in inhibiting both 14-3-3 protein processing and DNA fragmentation, we still observed considerable discrepancy between the concentrations required for caspase inhibition (0.5 µM) on one hand and for significant effects on these processes (50 µM) on the other hand (data not shown).
DISCUSSION
- (a)
We report that the caspase inhibitor zDEVD-fmk at 0.5 µM or more inhibits the cleavage of three different caspase substrates (Figs. 5 and 6B). Importantly, we have previously shown by anti-caspase-3 immunoblotting that zDEVD-fmk (as well as zVAD-fmk) actually covalently binds to caspase-3 under our experimental conditions [Kuželová et al., 2007]. The slight shift of the band corresponding to caspase-3 towards higher MW (cf. also lanes 2 and 7 vs. lanes 3–6 and 8 in Fig. 6C of the present work) indicates that the inhibitor was covalently attached to apparently all caspase-3 molecules when added at up from 0.5 µM concentration. Up from 1 µM concentration, the inhibitor also impedes pro-caspase autoprocessing (additional band at about 20 kDa is formed in anti-caspase-3 immunoblots, see also Fig. 6C, lanes 3–6 and 8). Besides caspase-3, zDEVD-fmk is also known to potently block the activity of caspases-6, -7, -8, and -10 [Ekert et al., 1999]. On the other hand, this inhibitor produces only partial effects on 14-3-3 processing even at 50 µM concentration (Fig. 6A).
- (b)
Further circumstantial confirmation of an efficient caspase-3 inhibition by zDEVD-fmk comes from the fact that 5 µM zDEVD-fmk fully prevents the cleavage of 14-3-3 γ. Despite it, the inhibitor is not able to reduce the cleavage of 14-3-3 τ and has only limited effect on the cleavage of 14-3-3 β, ε, and ζ (Fig. 6A).
- (c)
The pan-caspase inhibitor zVAD-fmk is markedly more efficient than zDEVD-fmk in preventing both the overall decrease in 14-3-3 expression level and the fragment formation. Following general recommendations, the effective final concentration of zVAD-fmk should be 5–20 µM. In our experimental system, the concentration of 2 µM was sufficient to completely inhibit the DEVDase activity [Kuželová et al., 2007]. However, a markedly higher concentration (50 µM) was required to achieve significant effects on 14-3-3 processing (Fig. 6A).
Altogether, these results clearly show that while caspase-3 probably contributes to 14-3-3 cleavage (especially in the case of 14-3-3 γ and ε), other mechanism has to be involved in this process. Although the selectivity of short peptide-based caspase substrates with regard to the individual caspases is limited [McStay et al., 2008], the data presented in Figure 5 indicate that caspases 9 and 2 are not active in the presence of 5 µM zDEVD-fmk. It is important to realize that the specificity of zVAD-fmk for caspases is compromised when the inhibitor is used at high concentrations (such as 50 µM or more) [Schotte et al., 1999; Rozman-Pungercar et al., 2003], so that the inhibitory effect of zVAD-fmk on both 14-3-3 processing and the apoptotic DNA fragmentation (Figs. 6 and 7) is not necessarily mediated by its effects on the caspase family. With regard to the known in vitro affinity of the used caspase inhibitors for different caspases [Ekert et al., 1999] we believe that none of them can be responsible for the observed 14-3-3 τ processing. We have recently shown that the fluorescently labeled caspase inhibitor FAM-DEVD-fmk binds not only to caspase-3 but also to another target in apoptotic JURL-MK1 cells [Kuželová et al., 2007]. The corresponding strong fluorescent signal can be inhibited by zVAD-fmk at high concentration (50 µM) but not by zDEVD-fmk. Thus, a protease with substrate specificity similar to that of caspase-3 may operate in apoptotic cells and be responsible for 14-3-3 cleavage. Possible candidates are cathepsins, non-cysteine proteases cleaving after Asp residue. However, we failed to detect any effect of pepstatin A (up to 7.5 µM, data not shown), which inhibits cathepsin activity, on the cleavage of 14-3-3 proteins.
The changes in the expression levels of all 14-3-3 isoforms can be largely prevented under the experimental conditions which lead to the inhibition of DNA fragmentation (Fig. 7). It thus appears that the decrease of 14-3-3 expression and cleavage of some 14-3-3 isoforms are related to the apoptosis progression and may represent an integral part of the programmed cell death or at least of some apoptotic pathways.
The MW of 14-3-3 fragments which are produced during the apoptosis is only slightly lower in comparison with the full-length proteins and the cleavage thus corresponds to the removal of several amino acids only. If the truncation took place at the N-terminus of the protein, it would probably negatively affect the ability of 14-3-3's to form dimers and thereby the affinity for client proteins. However, we judge from indirect indications that 14-3-3 fragments rather lack C-terminal amino acids: the fragment of 14-3-3 β is recognized by the antibody specific for the N-terminus of the protein (sc-1657) but not by C-terminal anti-14-3-3 β antibody (sc-25276) (Fig. 4). Similarly, the antibody against the C-terminus of 14-3-3 ε (AB 9732) detects solely the full-length protein. The series of peptides identified by MALDI from the spot presumably corresponding to the cleaved 14-3-3 τ contains peptides from the N-terminus while it lacks the C-terminal peptide which is present in the intact protein (Fig. 3B). Moreover, 14-3-3 ε possesses additional amino acids at the C-terminus in comparison with the other isoforms (Fig. 4, top) and this would explain the fact that the lost sequence of 14-3-3 ε is larger than that of 14-3-3 β, γ, ζ, or τ (the shift towards lower MW is larger in the case of 14-3-3 ε). The isoform ε displays also the highest shift in pI on 2D gels (Fig. 3A). It follows from the sequence alignment (top of Fig. 4) that the loss of 6–7 amino acids from the C-terminal tail of 14-3-3 β or τ compared to 17 amino acids from that of 14-3-3 ε would result in loss of 2 compared to 6 acidic charges. In the case of γ isoform, the shift in pI is larger than that for β and τ (Fig. 3A) suggesting that three acidic charges (eight amino acids from the C-terminus) might be removed.
Nomura et al. 2003 have shown using immunoprecipitation that the removal of seven C-terminal amino acids from 14-3-3 τ results in the loss of the affinity for Bax. Similarly, C-terminal cleavage of 14-3-3 ε has been shown to impair the ability of the protein to bind Bad and the expression of the truncated 14-3-3 ε in 293T cells enhanced Bad-mediated apoptosis [Won et al., 2003]. It is thus reasonable to suppose that 14-3-3 cleavage leads to the release of the proapoptotic client proteins and forms a positive feedback allowing for amplification of proapoptotic signals. It is also possible that the truncated forms of 14-3-3 proteins have an additional role in the apoptosis progression. The C-terminal tail of 14-3-3 γ was recently shown to be required for a full and specific interaction with the scaffold protein KSR1 which regulates signal transduction through Raf/MEK/ERK pathway [Jagemann et al., 2008]. It is interesting to note in this regard that the truncation of 14-3-3 ζ has been reported to enhance its affinity for Raf-1 and Bad [Truong et al., 2002]. A study of this isoform has indicated that the C-terminal tail of the protein in ligand-free form occupies the ligand-binding groove and inhibits, to some extent, inappropriate associations [Silhan et al., 2004]. However, we observed only weak cleavage of 14-3-3 ζ isoform during Imatinib-induced apoptosis of JURL-MK1 cells and no cleavage of this particular isoform was found by Nomura et al. after etoposide treatment of HeLa cells.
In conclusion, we believe that the decrease of 14-3-3 protein expression level and the removal of the C-terminus belong to common processes contributing to the apoptosis regulation, regardless of the cell type and of the inducing stimulus. Our results show that the cleavage of 14-3-3 proteins is executed by at least two different mechanisms probably involving caspase-3 and still unknown protease beyond the caspase family, in an isoform-specific manner.
Acknowledgements
We acknowledge the expert technical assistance of J. Sedlmaierová and H. Pilcová.




