The nucleosomal binding protein NSBP1 is highly expressed in the placenta and modulates the expression of differentiation markers in placental Rcho-1 cells†
Hitoshi Shirakawa, Mark Rochman, and Takashi Furusawa contributed equally to this work.
Abstract
We report that NSBP1, a nucleosome binding protein that affects the structure of chromatin, is highly expressed in mouse placenta. In Rcho-1 cells, which recapitulate the differentiation of trophoblast giant cells of living placenta, NSBP1 expression is linked to differentiation. Disregulation of NSBP1 protein levels, by either siRNA treatment or by overexpression, alters the expression of several members of the prolactin gene family without affecting the levels of several transcription factors involved in placental differentiation. Our studies identify NSBP1 as a nucleosome binding protein that modulates the expression of prolactin gene family members most likely by inducing changes in chromatin structure. J. Cell. Biochem. 106: 651–658, 2009. © 2009 Wiley-Liss, Inc.
Proper development and differentiation is contingent on the correct execution of a preprogrammed, orderly process that involves multiple, highly regulated, changes in gene expression. Given the central role of chromatin structure in regulating gene expression, it can be expected that structural chromatin-binding proteins such as members of the high mobility group (HMG) protein superfamily, could affect gene expression during differentiation [Reeves, 2001; Sgarra et al., 2004; Bianchi and Agresti, 2005; Hock et al., 2007].
The high mobility group N (HMGN) proteins, a major family of the HMG superfamily, are unique among the numerous proteins that target chromatin in that they are the only nuclear proteins shown to specifically bind to the 147 base pair nucleosome core particle, the building block of the chromatin fiber [Bustin, 2001]. Studies with in vitro chromatin reconstitution systems, with cells grown in tissue culture, and with genetically altered mice revealed that misregulation of HMGN expression affects transcription and developmental processes [Bustin, 2001; Hock et al., 2007].
We have previously described a gene coding for a novel protein named NSBP1 (Nucleosome Binding Protein 1, originally named NBP-45), which is structurally related to HMGNs but is not a typical HMGN protein [Shirakawa et al., 2000]. NSBP1 binds specifically to nucleosomes and can stimulate transcription from a reporter gene; however, the cellular function of this protein is not yet known. Our initial analyses revealed that NSBP1 transcripts are especially abundant in 7-day-old mice embryos, suggesting a role for the protein in mouse embryogenesis [Shirakawa et al., 2000]. We now report that the protein is highly expressed in placental rather than embryonic tissues, and is highly enriched in trophoblast giant cells, which play a key role in placental differentiation [Simmons and Cross, 2005; Simmons et al., 2008]. To gain insights into its possible function in placental development, we examined its role in regulating the expression prolactin gene family members in Rcho-1 cells, a cell line that serves as a model system for studies on the development of trophoblast giant cells [Hamlin et al., 1994]. In these cells the expression of specific lactogens is linked to differentiation in a manner similar to their expression in living placenta. We discovered that altered levels of NSBP1 protein in Rcho-1 cells affected the expression pattern of several prolacting genes and placental lactogens. Our studies identify NSBP1 as a nucleosome binding protein that modulates the transcriptional fidelity of trophoblast cells.
MATERIALS AND METHODS
Cell Culture and Generation of Stable Clones
Rat choriocarcinoma cells Rcho-1 [Sahgal et al., 2006], a gift from Dr. M.J. Soares, University of Kansas Medical Center, Kansas City, USA, were maintained in NCTC-135 medium (Sigma) or RPMI (Invitrogen) supplemented with 20% heat inactivated fetal bovine serum (FBS, ICN), 10 mM HEPES, 50 µM 2-mercaptoethanol, 1 mM sodium pyruvate, 100 U/ml of penicillin, and 100 µg/ml of streptomycin. Differentiation of cells was induced by growing the cells to near confluence in FBS supplemented culture medium and then replacing the growth medium with differentiation medium (DM) which is growing medium supplemented with either 1% or 10% heat inactivated horse serum (GIBCO). For preparation of stable Rcho-1 clones, undifferentiated Rcho-1 cells were transfected with plasmids expressing wild NSBP1-GFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Selection was performed with 250 µg/ml of G418 for 2 weeks and then GFP positive cells were collected by FACS sorting and split into 96-well plate, 30 cells per well. Following additional 2 week of selection, GFP-positive stably transfected clones were expanded and used for experiments. Stable clones were routinely maintained in medium containing 0.1 µg/ml of G418. H4IIE, a rat hepatoma cell line (ATCC) was cultured in DMEM + 10% FBS.
In Situ Hybridization
Sense- or antisense RNA probes for in situ hybridization of E9.5 to E11.5 were generated by transcribing the plasmid containing full-length of mouse Nsbp1 cDNA in pCR-BluntIITOPO [Shirakawa et al., 2000] with either T7 or SP6 RNA polymerases in the presence of 35S-labeled nucleotides. The image of the radioactive signal produced in the emulsion was observed with dark field illumination. The Nsbp1 transcript in E7.5 embryos were visualized by whole mount in situ hybridization using probes containing digoxigenin-11-UTP (Roche Diagnostics, Germany) which were visualized using alkaline phosphatase labeled anti Digoxigenin (Roche Diagnostics) as recommended by the manufacturer.
Mice
C57BL/6N mice, 8–16 weeks old, were obtained from the NIH animal facility. For pregnancy dating, the finding of the vaginal plug was taken as evidence of copulation and for dating the onset of pregnancy.
Northern Hybridization, Antibodies, Immunofluorescence, and Western Blot Analysis
Total RNA was isolated from tissues and cell lines using Isogen (Nippon Gene Co., Japan), electrophoresed in 1.5% formaldehyde-agarose gel, transferred to Hybond-N+ nylon membranes (Amersham) and probed with 32P-labeled probes. Affinity purified antibodies to HMGN1 and HMGN2 were as described before [Postnikov et al., 1995]. Antibodies to mouse NSBP1 were prepared by immunizing rabbits with recombinant NSBP1 [Shirakawa et al., 2000]. The antibodies did not react with HMGN1 or HMGN2. Western [Shirakawa et al., 2000] and immunofluorescence [Furusawa et al., 2006] analyses were done as described.
Small Interfering RNA
For prevention of NSBP1 expression during differentiation, subconfluent, undifferentiated Rcho-1 cells were transfected with siNsbp1 ON-TARGET plus SMARTpool (Dharmacon catalogue L-044143-00) which consists of a set of four oligonucleotides targeting the open reading frame of Nsbp1, using Dharmafect 4 transfection reagent (Dharmacon). After the cells reached confluence (approximately 48 h post-transfection), differentiation was induced by changing the medium to DM (differentiation medium) and cells were retransfected with siNsbp1 mix. DM was changed every 24 h and cells were collected 72 hrs after induction of differentiation. For controls, Rcho-1 cells were transfected with control complimentary oligonucleotides from Dharmacon (ON-TARGETplus, Non-targeting siRNA, D-001810-01-05). For down regulation of NSBP1 in differentiated Rcho-1 cells, 21 base pair double stranded siRNA directed against Nsbp1 transcripts (nucleotides 86–106 in the cDNA sequence) was produced by annealing of complementary oligonucleotides in annealing buffer (10 mM HEPES–NaOH, 150 mM NaCl, pH7). Double stranded RNA was transfected into differentiated cells using Lipofectamine 2000 (Invitrogen). Control cells were transfected with the also following double stranded RNA purchased from Dharmacon: (CAGUCGCGUUUGCGACUGGdTdT/dTdTGUCAGCGCAAACGCUGACC). The medium containing liposome–RNA complex was changed freshly every 48 h. After 2 or 7 days, cells were harvested and total RNA was isolated and used as a template for cDNA synthesis. siRNA mix and oligonucleotides were purchased from Dharmacon Research Inc. (Lafayette, CO).
Quantitative RT-PCR
Total RNA was prepared by Trizol isolation and further purified by RNeasy kit (Qiagene). 0.5–1 µg of RNA was used as a template for cDNA synthesis. cDNA was prepared by iScript (Biorad). Equal aliquots of cDNA were used as a template for quantitative PCR using Applied Biosystems Sequence Detection System 7000 (Foster City, CA). The gene specific primers are listed in Supplemental Table S1.
Chip Analysis
The chromatin immunoprecipitation (ChIP) assay based on the methods of Farnham and colleagues (http://www.genomecenter.ucdavis.edu/farnham/protocols/tissues.html) was conducted by using a kit from Upstate Biotechnology (Lake Placid, NY). The immunoprecipitated DNAs by rabbit anti-mouse Nsbp1 or normal rabbit IgG, as negative control, were used as the template for PCR that was performed by using specific primers for the promoter region of the rat PL-1, d/t PRP or β-actin listed in Supplemental Table S1. Amplified DNA fragments were electrophoresed on 1.5% agarose or 10% polyacrylamide gel and visualized by ethidium bromide staining.
RESULTS AND DISCUSSION
High NSBP1 Expression in Placenta and in Trophoblast Giant Cells
Our previous analysis suggested that NSBP1 transcripts were especially abundant in 7.5-day-old embryos [Shirakawa et al., 2000], raising the possibility that the protein may play a role in embryonic development. Therefore, to gain insights into the cellular function of NSBP1, we first examined its expression pattern in E7.5 embryos. Surprisingly, whole mount in situ hybridization and immunofluorescence analysis reveal that at E7.5 NSBP1 transcripts and protein are detectable only in the ectoplacental cone, an extraembryonic tissue composed mainly of diploid trophoblast cells, which develops into placenta (Fig. 1A). At E9.5 and E11.5, strong Nsbp1 transcript expression is detected in the giant trophoblast, spongiotrophoblast and decidual cells of the placenta (Fig. 1B) while the expression in the developing embryo remains relatively weak. Immunofluorescence analyses verified that the NSBP1 protein is strongly expressed in these regions (Fig. 1C,D). The expression of the protein is especially prominent in the Trophoblast giant cells, in spongiotrophoblasts, and in trophoblasts located in the placental labyrinth (Fig. 1D,E).
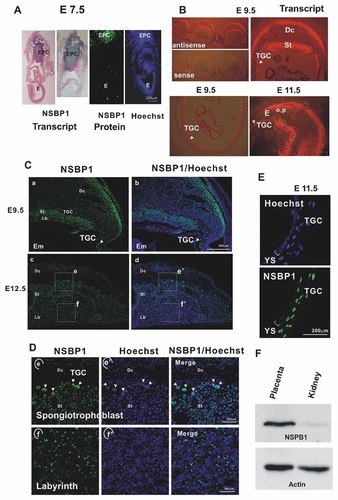
Expression of NSBP1 during mouse embryonic development. A: Whole mount in situ hybridization and immunofluorescence analysis of NSBP1 expression in 7.5-day-old (E7.5) mouse embryo reveals prominent NSBP1 expression in the ectoplacental cone (EPC) but not in embryonic tissues (E). B: In situ hybridization analysis of Nsbp1 expression in E9.5 and E11.5 embryos indicates strong NSBP1 expression is spongiotrophoblasts (St), trophoblast giant cells (TGC) and in outermost layers of the decidua (Dc). Upper left panels demonstrate that positive hybridization signals are obtained in E9.5 embryos only with the antisense probe. C: Immunofluorescence analyses of NSBP1 in E9.5 and E12.5 placenta. Protein expression is detected in decidual cells (Dc) in trophoplast giant cells (TGC) in the spongiothrophoblasts (St) and in the large trophoblast cells of the labyrinth region (Lb). D: High magnification of the areas selected from panel C. Note high NSBP1 expression in trophoblast giant cells (arrows, TGC) and in the large cells of the labyrinth (panels f and f′). E: NSBP1 expression in giant cells of the trophoblast layer at E11.5 YS: yolk sack. F: Western blot analyses of whole tissue extracts prepared from the mouse E11 placenta and from adult mouse kidney indicate high expression of NSBP1 in placenta.
Western analyses of extracts prepared from day 11 placenta and from adult kidney also indicate that the expression of NSBP1 in placenta is higher than that in a fully differentiated organ such as the kidney (Fig. 1F). The prominent expression of NSBP1 in trophoblasts suggests a role for the protein in their differentiation or function.
Developmentally Regulated Nsbp1 Expression in Placental Rcho-1 Cells
We examined the possible role of NSBP1 in trophoblast giant cells by analyzing the expression of the protein in Rcho-1 trophoblast cells which serve as a model system for placental development since they can be maintained in a proliferative state, or induced to differentiate into cells undergoing endoreduplication that morphologically and functionally resemble in vivo developing trophoblast giant cells [Hamlin et al., 1994].
Undifferentiated cells, cultured to confluence in growth medium which contained 20% fetal bovine serum (see Materials and Methods Section) can be induced to differentiate into trophoblast giant cells by switching to a medium supplemented with horse serum [Hamlin et al., 1994; Sahgal et al., 2006]. Northern analyses reveals that the levels of Nspb1 transcripts in fully differentiated Rcho-1 cells are significantly higher than in undifferentiated cells (Fig. 2A, Northern blot) and quantitative RT-PCR reveals a gradual increase in the Nsbp1 transcripts during differentiation (Fig. 2A, RT PCR) suggesting a link between increased Nsbp1 transcription and trophoblast differentiation. The levels of transcripts coding for the nucleosomal binding protein HMGN1, a protein that is related to NSBP1 and served as a control [Shirakawa et al., 2000], were not linked to Rcho-1 differentiation. In agreement with the RNA analyses, the levels of NSBP1 protein, but not those of HMGN1, increased during differentiation (Fig. 2A, Western blot). Observation of DAPI-stained undifferentiated cultures reveals the presence of a few giant cells (Fig. 2Bb) and immunofluorescence analyses reveal that only these giant cells contain NSBP1 (Fig. 2Bc). In the differentiated cultures that containing mainly giant cells, most cells also express NSBP1 (Fig. 2Be). Thus, the expression of NSBP1 is linked to the differentiation of Rcho-1 cells.
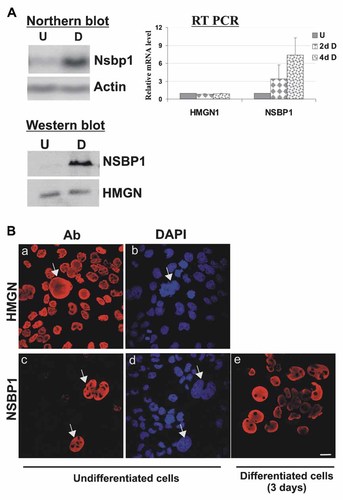
Differentiation-dependent expression of NSBP1 in Rcho-1 cells. A: Northern blot, quantitative RT-PCR, and Western blot analyses demonstrate induction of Nsbp1 expression in differentiated Rcho-1 cells. U, undifferentiated cells; D, differentiated cells, 2d and 4d indicate days after induction of differentiation. In the western blots analyses HMGN serves as a loading control. B: Immunofluorescence analyses of NSBP1 and HMGN expression in undifferentiated (a–d) and differentiated (e) cells. Note that the few giant cells present in undifferentiated cultures express NSBP1 (arrows in c and d) while the smaller undifferentiated cells do not. The related nuclear protein HMGN1 is uniformly expressed in all cells (panel a).
NSBP1 Modulates Prolactin Gene Family Expression in Rcho-1 Cells
The differentiation of Rcho-1 cells in culture recapitulates the in vivo differentiation of trophoblast giant cells and is characterized by the sequential expression of members of the prolactin gene family which also serve as differentiation markers [Hamlin et al., 1994]. Since NSBP1 is abundant in placental cells (Fig. 1) and since its expression in cells is linked to differentiation (Fig. 2) we tested whether NSBP1 affects the differentiation process by analyzing the expression of these genes following alterations in NSBP1 level.
To test whether the presence of NSBP1 is required for the maintenance of the differentiation state, we used siRNA to down regulate the levels of Nspb1 in fully differentiated Rcho-1 cells. Following siRNA treatment, the levels of Nsbp1 transcripts were only 20% of that found in cells transfected with control siRNA (Fig. 3A Q-PCR). Consistent with decreased mRNA levels, within 7 days of treatment, the levels of NSBP1 protein were reduced by more than 90% (Fig. 3A, Westerns). The siRNA targeted specifically Nsbp1 transcripts since the levels of HMGN2 and actin were not affected. Down regulation of NSBP1 in differentiated cells did not lead to detectable morphological changes (not shown) and did not affect significantly the mRNA levels of differentiation associated transcription factors such as Hand1, GATA2, GATA3 or metalloproteinase 9 (Fig. 3B). Nevertheless, loss of NSBP1 down regulated the expression levels of several trophoblast specific PL transcripts such as PL-1 and PLP-A which serve as early and late markers of trophoblast differentiation, respectively (Fig. 3B). The results suggest that NSBP1 facilitates the maintenance of PL expression in differentiated Rcho-1 cells.
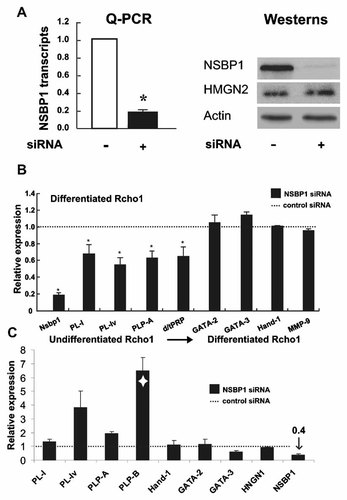
Down regulation of NSBP1 expression alters the expression of PL genes. A: Quantitative RT-PCR (Q-PCR) and Western blot analysis demonstrate that siRNA treatment down regulates the expression of NSBP1 in differentiated Rcho-1 cells. The siRNA treatment commenced 4 days after the onset of differentiation by changing the growth medium. B: In differentiated Rcho-1 cells depletion of NSBP1 leads to down regulation of placental lactogens and members of the prolactin gene family. The levels of transcripts were quantified by RT-PCR and normalized to glyceraldeyde-3-phosphate dehydrogenase (GAPDH). The normalized value of the various transcripts in the cells transfected with control siRNA was set to 1.0 (dotted line). * Indicates statistical significant difference, as determined by the Student's t-test. Note that the expression of transcriptional regulators was not affected. C: Aberrant expression of placental lactogens in Rcho-1 cells depleted of NSBP1 prior to, and during differentiation. Shown are the relative levels of placental lactogens in Rcho-1 cells after 72 h of induction of differentiation. The normalized value of the various transcripts in differentiated cells transfected with control siRNA was set to 1.0 (dotted line). White diamond indicates a PLP-B transcript, a marker of spongiotrophoblast cells. Mean value of three independent experiments is shown.
To test whether NSBP1 expression is necessary for transition from undifferentiated to differentiated stage, we cultured undifferentiated Rcho-1 cells in the presence of Nsbp1-specific siRNA for 48 h and then induced them to differentiate, still in the presence of this siRNA (Fig. 3C). Quantitative RT-PCR analyses of RNA extracted 72 h after differentiation indicated that the siRNA treatment efficiently prevented increase in mRNA of NSBP1 by approximately 60%, as compared to control siRNA treated cultures. Depletion of NSBP1 induced the early expression of PL-Iv and PLP-A, which are late differentiation markers and also dramatically stimulated PLP-B transcription, a marker of spongiotrophoblast differentiation, which normally is expressed at very low levels in trophoblast giant cells. Thus, depletion of NSBP1 during differentiation altered the transcription profile of the differentiating Rcho-1 cells. Similarly to the down regulation of NSBP1 in differentiated cells (Fig. 3B), depletion of NSBP1did not affect significantly the expression of transcriptional regulators that modulate differentiation such as Hand1, GATA2 and GATA3. In agreement, these siRNA treated cells were able to differentiate into trophoblast giant cells. Thus, depletion of NSBP1 affected specifically the expression of prolactin-related genes without affecting the expression of transcriptional regulators and without noticeably affecting the differentiation of Rcho-1 cells. The expression profile of the prolactin gene family is related to cellular differentiation therefore it is not surprising that NSBP1 affects gene expression in a differentiation specific manner.
To assess whether elevation of NSBP1 levels would affect differentiation we established Rcho-1 cells stably overexpressing the NSBP1 protein. In the undifferentiated cells, overexpression of NSBP1, elevated the transcription of PL-1, an early marker of placental differentiation, without altering the expression of transcription factors such a GATA3 or Hand1 (Fig. 4A) and without affecting the cellular morphology (not shown). Thus, down regulation of NSBP1 in differentiated cells reduces the levels of PL-1 (Fig. 3B) while upregulation of NSBP1 in undifferentiated cells upregulates PL-1 (Fig. 4A). The results suggest that NSBP1 regulates the expression fidelity of PL-1, a lactogen expressed during the early stages of trophoblast differentiation.
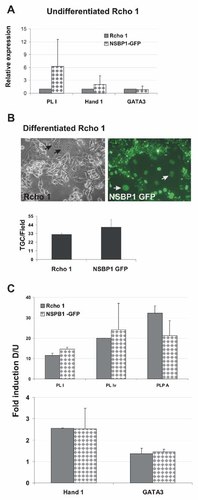
Upregulation of NSBP1 affects PL-1 expression but not Rcho-1 differentiation. A: Stable overexpression of NSBP1-GFP in undifferentiated Rcho-1 cells elevates the levels of PL-1. The levels of the PL-1, Hand1 and GATA3 transcripts in the stably transfected cells, determined by quantitative RT-PCR and normalized GAPDH, are compared to those of untransfected cells, which are set to 1.0. B: Overexpression of NSBP1-GFP does not affect differentiation. Phase contrast and fluorescence images of cells differentiated for 72 h, indicate that the majority of cells express the NSBP1-GFP protein. The bar graph below the images indicates that overexpression of the protein did not significantly alter the average number of trophoblast giant cells (TGC) per field. Three fields from cells transfected with two different clones were counted. C: Forced expression of NSBP1-GFP does not affect significantly the expression of PL genes following differentiation. Shown are the levels of the various transcripts (indicated below the columns) relative to those in undifferentiated cells. The data represent the mean of two independent clones.
To test whether elevated levels of NSBP1 affects the differentiation, we analyzed the ability of the stably transfected cells to undergo differentiation. After transfection and 2 weeks selection, the undifferentiated cells expressing NSBP1-GFP were isolated by FACS, plated, propagated and induced to differentiate. After 72 h of differentiation, the cells overexpressing NSBP1 differentiated to the same extent as the control cells, as judged by the overall appearance of the culture and by counting the number of giant cells in the culture (Fig. 4B). As judged from the fluorescence intensity, the small undifferentiated cells expressed NSBP-1 at level comparable to the differentiated large cells. thus overexpression of NSBP1 did not have major effects on the differentiation process itself. Moreover, in the differentiated cells, exogenous expression of NSBP1 did not affect the expression of several PL genes (Fig. 4C), perhaps because these cells already contain sufficient levels of endogenous protein to adequately regulate the PL genes.
While the expression pattern of PL genes in human and rodent placenta is well elucidated, the mechanisms regulating their expression are not fully characterized. Since NSBP1 is a nucleosome binding protein, we explored the possibility that it binds to the chromatin of the placental lactogens. To this end we performed chromatin immunoprecipitation (ChIP) experiments in Rcho-1 and H4IIE cells. Both cells contain NSBP1 (Fig. 5A) but only Rcho-1 cells express prolactin genes. The results indicate that only in Rcho-1 cells, NSBP1 is associated with the chromatin of PL-1 and d/tPRP, two lactogens whose expression is affected by NSBP1 (Fig. 5B).
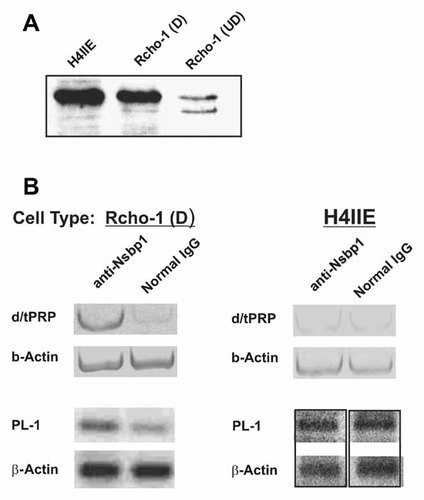
Cell specific association of NSBP1 is with the chromatin of PL genes. A: High expression of NSBP1 in H4IIE and in differentiated Rcho-1 cells. Equal amounts of cell extracts were added to each lane. B: ChIP analyses done with anti-NSBP1 demonstrate association of NSBP1 with the chromatin of PL-1 or d/tPRP in Rcho-1 cells but not in H4IIE cells.
In conclusion, we find that NSBP1 is highly expressed in trophoblasts, that its expression is related to differentiation and that it interacts with the chromatin of members of the prolacting gene family, in a cell type specific manner. Taken together with our finding that NSBP1 affect the expression of prolactin-related genes without affecting the levels of several transcriptional regulators, and with previous finding that mouse PL-1 and PL-2 genes are regulated by GATA2 or GATA3, and Hand1 [Ma et al., 1997] the results suggest that that NSBP1 modulates the expression of the placental lactogens by inducing changes in chromatin structure rather than by affecting the levels of specific transcription factors. Since the expression of placental lactogens in Rcho-1 reflect trophoblast differentiation in vivo [Simmons et al., 2008] it is possible that NSBP1 may affect placental differentiation.
Acknowledgements
This work was partially supported by Japan Society for the Promotion of Science (The US-Japan cooperative cancer research program) to HS and by the intramural research program of the National Cancer Institute, NIH. We thank Dr. M.J. Soares, Institute of Maternal-Fetal Biology, University of Kansas Medical Center, Kansas City, for a gift of Rcho-1 cells.




