Parathyroid hormone regulation of the human bone sialoprotein gene transcription is mediated through two cAMP response elements
Abstract
Parathyroid hormone (PTH) regulates serum calcium and inorganic phosphate levels through its actions on kidney and bone. Bone sialoprotein (BSP) is an early marker of osteoblast differentiation and bone metabolism. We here report that two cAMP response elements (CRE) in the human BSP gene promoter are target of PTH. In human osteoblast-like Saos2 cells, PTH (human 1−34 PTH, 10 nM) increased BSP mRNA and protein levels at 3 h. From transient transfection assays, 2- to 2.5-fold increase in transcription by PTH was observed at 3 and 6 h in −184, −211, −428, −868, and −927 luciferase constructs that included the human BSP gene promoter. Effect of PTH was abrogated by 2 bp mutations in either the CRE1 (−79 to −72) or CRE2 (−674 to −667). Luciferase activities induced by PTH were blocked by protein kinase A inhibitor H89 and tyrosine kinase inhibitor herbimycin A. Gel shift analyses showed that PTH increased binding of nuclear proteins to the CRE1 and CRE2 elements. The CRE1–protein and CRE2–protein complexes were disrupted by CRE binding protein 1 (CREB1) antibodies and supershifted by phospho-CREB1 antibody. ChIP assays detected binding of CREB1 and phospho-CREB1 to a chromatin fragment containing CRE1 and CRE2, and increased binding of phospho-CREB1 to the both sites. These studies demonstrate that PTH stimulates human BSP gene transcription by targeting the two CREs in the promoter of the human BSP gene. J. Cell. Biochem. 106: 618–625, 2009. © 2009 Wiley-Liss, Inc.
Abbreviations used:
BSP, bone sialoprotein; ChIP, chromatin immunoprecipitation; FGF2, fibroblast growth factor 2; FRE, FGF response element; bp, base pair(s); nts, nucleotides; PKC, protein kinase C; PKA, cAMP dependent protein kinase; Pit-1, pituitary-specific transcription factor-1; LUC, luciferase; AP1, activator protein 1; NFκB, nuclear factor-kappa B; CRE, cAMP response element; CREB, CRE binding protein; α-MEM, α-minimum essential medium; Runx2, runt homeodomain protein 2.
Parathyroid hormone (PTH) is a peptide hormone functioning as a major mediator of bone remodeling and as an essential regulator of calcium homeostasis [Strewler et al., 1987]. These activities of PTH reside within the first 34 residues of the 84 amino acid hormone [Hilliker et al., 1996]. The primary sites of action of PTH are considered to be kidney and bone. Very small decrements in serum calcium levels induce the secretion of PTH from the parathyroid glands initiating a rapid response to raise serum calcium levels by acting directly on kidney and bone or indirectly on intestine facilitating calcium absorption [Potts et al., 1997; Silverberg et al., 1999]. In bone, PTH has both catabolic and anabolic effects that depend on the mode of administration [Tregear et al., 1973]. Continuous administration of PTH in the same doses causes catabolic skeletal effects, whereas intermittent (subcutaneous or intraperitoneal) injection of PTH once a day stimulates new bone formation and increased bone mass in patients with osteoporosis [Sone et al., 1995; Lindsay et al., 1997; Cosman and Lindsay, 1998] and ovariectomized monkeys [Brommage et al., 1999]. This increase in bone mass may result from an increase in the number of osteoblasts either by formation of new osteoblasts from bone lining cells [Dobnbig and Turner, 1995] and/or by stimulating proliferation of mature osteoblasts [Partridge et al., 1985].
PTH receptors are found in osteoblasts, chondrocytes and renal epithelial cells and osteoclasts do not possess PTH receptors [Luben et al., 1976; Partridge et al., 1981; Majeska and Rodan, 1982; Suda et al., 1996]. The receptor for PTH belongs to the superfamily of the seven transmembrane G protein coupled receptors [Abou-Samra et al., 1992; Stewart, 1996]. As a consequence of PTH binding to its receptors, the Gs proteins mediate the activation of adenylate cyclase, which leads to elevate cyclic AMP (cAMP) levels in the cells and activates cAMP dependent protein kinase A (PKA). PTH also activates phospholipase C (PLC), leading to the formation of two second messengers. Diacylglycerol which promotes translocation and activation of protein kinase C (PKC) and inositol 1,4,5-trisphosphate (1,4,5-IP3) which induces Ca2+ release from intracellular Ca2+ pool [Majeska et al., 1980; Bidwell et al., 1991a,b; Berridge, 1993; Ahlström and Lamberg-Allardt, 1997; Kondo et al., 1997].
Bone sialoprotein (BSP) is phosphorylated and sulfated glycoprotein that represents one of the major noncollagenous, extracellular matrix proteins associated with mineralized tissues [Fisher et al., 1990; Ganss et al., 1999; Ogata, 2008]. High BSP expression coincides with de novo bone formation [Chen et al., 1992]. BSP is primarily expressed by mature osteoblasts, osteoclasts and hypertrophic chondrocytes [Bianco et al., 1991]. And also BSP is expressed by breast, prostate and lung cancers and to be associated with the formation of ectopic hydroxyapatite microcrystals in the tumor tissues [Waltregny et al., 2000; Ogata, 2008]. The rat, mouse, and human BSP genes have been cloned and partially characterized [Kerr et al., 1993; Li and Sodek, 1993; Kim et al., 1994; Benson et al., 1999; Kiyoshima et al., 2002]. Human BSP gene promoter have an inverted TATA box (nt −28 to −23) [Kim et al., 1994], an inverted CCAAT box (nt −54 to −50) which required for basal transcription [Kim and Sodek, 1999; Kiyoshima et al., 2002] and two cAMP response elements (CRE1; −79 to −72 and CRE2; −674 to −667) [Huang et al., 2005]. In addition, a FGF2 response element (−96 to −89) [Shimizu-Sasaki et al., 2001; Nakayama et al., 2006; Detry et al., 2008], three activating protein 1 response elements (AP1(1); −148 to −142, AP1(2); −483 to −477 and AP1(3); −797 to −791) [Kim et al., 1994], and a homeodomein protein-binding site (HOX; −200 to −191) [Kim et al., 1994; Nakayama et al., 2006] have been characterized.
We previously reported that PTH increased rat BSP gene transcription by targeting a pituitary-specific transcription factor-1 (Pit-1) motif in the promoter of the rat BSP gene [Ogata et al., 2000]. To elucidate the molecular mechanism of the PTH regulate human BSP gene transcription, we have analyzed the effects of PTH on the expression of human BSP in human osteoblast-like Saos2 cells. These studies have revealed two CRE sites mediate PTH-induced human BSP gene expression in osteoblastic cells.
METHODS
Materials
Alpha-minimum essential medium (α-MEM), fetal calf serum (FCS), lipofectamine, penicillin and streptomycin, trypsin–EDTA were obtained from Invitrogen (Carlsbad, CA). The pGL3-basic and pSV-β-galactosidase (β-Gal) control vectors were purchased from Promega Co. (Madison, WI). The protein kinase inhibitors H89 and H7 were from Seikagaku Corporation (Tokyo, Japan). Tyrosine kinase inhibitor herbimycin A (HA) were purchased from WAKO Pure Chemical Industries, Ltd (Tokyo, Japan). EXScript RT reagent Kit and SYBR Premix Ex Taq were purchased from Takara (Tokyo, Japan). Chip-IT™ Express Enzymatic kit was from Active Motif (Carlsbad, CA). Human 1–34 PTH was from the Peptide Institute (Osaka, Japan).
Cell Culture
Human osteosarcoma cell line, Saos2 cells were cultured at 37°C in 5% CO2, 95% air in α-MEM containing 10% FCS. Cells were grown to confluence in 60 mm cell culture dishes and then cultured in α-MEM without serum for 12 h and incubated with or without PTH (10 nM) for time periods extending from 3 to 24 h. Total RNA was isolated from triplicate cultures and analyzed for the expressions of BSP, Runx2 and Osterix mRNA by Northern blot and real-time PCR as described below.
Northern Blot
Total RNA from the Saos2 cells was extracted with guanidium thiocyanate and, following purification, 20 µg aliquots of RNA were fractionated on a 1.2% agarose gel and transferred onto a Hybond N+ membrane. Hybridizations were performed at 42°C with a 32P-labeled human BSP cDNA probe. Following hybridization, membrane was washed four times for 5 min each at 21°C in 300 mM sodium chloride, 30 mM trisodium citrate pH 7.0 containing 0.1% SDS. This was followed by two, 20 min washes at 55°C in 15 mM sodium chloride, 1.5 mM trisodium citrate pH 7.0, 0.1% SDS. The hybridized band, representing the human BSP mRNA, was scanned in a Bio-imaging analyzer (Fuji BAS 2000, Tokyo, Japan). Signals were quantitated by densitometry and normalized to the corresponding values for 18S RNA.
Western Blot
For Western blot analyses, cell lysate from Saos2 cells were separated on 10% SDS–PAGE and transferred onto a membrane. The membrane was then incubated for 3 h by anti-BSP polyclonal antibody (LF-100 was kindly provided by Dr. Larry W. Fisher) and anti-α tubulin antibody (sc-5286; Santa Cruz Biotechnology, Inc.). Anti-rabbit IgG conjugated with HRP (GE Healthcare UK, Ltd) was used as the secondary antibody. Immunoreactivities were detected by ECL plus Western Blotting Detection Reagents (GE Healthcare UK, Ltd).
Real-Time PCR
Total RNA (1 µg) was used as a template for cDNA synthesis. cDNA was prepared using EXScript RT Reagent Kit. Quantitative real-time PCR was performed using human BSP, Runx2, Osterix and GAPDH primer sets: BSP forward, 5′-CTGGCACAGGGTATACAGGGTTAG-3′; BSP reverse, 5′-ACTGGTGCCGTTTATGCCTTG-3′; Runx2 forward, 5′-ATGTGTGTTTGTTTCAGCAGCA-3′; Runx2 reverse, 5′-TCCCTAAAGTCACTCGGTATGTGTA-3′; Osterix forward, 5′-GCCAT TCTGGGCTTGGGTATC-3′; Osterix reverse, 5′-GAAGCCGGAGTGCAGGTATCA-3′; GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′; GAPDH reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′ using the SYBR Premix EX Taq in a TP800 thermal cycler dice real-time system (Takara). The amplification were performed in 25 µl of final volume containing 2x SYBR Premix EX Taq (12.5 µl), 0.2 µM forward and reverse primers (0.1 µl) and 25 ng cDNA (2.5 µl) for Runx2, Osterix and 10 ng (1.0 µl) cDNA for GAPDH. To reduce variability between replicates, PCR premixes, which contain all reagents except for cDNA, were prepared and aliquoted into 0.2 ml Hi-8-tubes (Takara). The thermal cycling conditions was 1 cycle at 95°C; 10 s, 40 cycles at 95°C; 5 s, 60°C; 30 s. Post-PCR melting curves confirmed the specificity of single-target amplification and fold expressions of Runx2 and Osterix relative to GAPDH were determined in triplicate.
Transient Transfection Assays
Exponentially growing Saos2 cells were used for transfection assays. Twenty-four hours after plating, the cells at 40–60% confluence were transfected using a lipofectamin reagent. The transfection mixture included 1 µg of a luciferase (LUC) construct and 2 µg pSV-β-galactosidase (β-gal) vectors as an internal control. Two days post-transfection, the cells were deprived of serum for 12 h, and PTH (10 nM) added for 3 or 6 h prior to harvesting. Various sized luciferase reporter constructs were prepared by ligating human BSP promoter DNA into pGL3-basic. Constructs −184Luc (−184 to +60), −211Luc (−211 to +60), −428LUC (−428 to +60), −868 LUC (−868 to +60), and −927 (−927 to +60) were prepared by PCR amplification. The amplified DNA was cloned in to the Bgl II site of the multiple cloning site of pGL3-basic [Kiyoshima et al., 2002]. The luciferase assay was performed according to the supplier's protocol using a luminescence reader Accuflex Lumi 400 to measure the luciferase activity. The protein kinase inhibitor H89 (5 µM) and H7 (5 µM) were used to inhibit protein kinase A and C. Herbimycin A (1 µM) was used for tyrosine kinase inhibition, respectively. Oligonucleotide-directed mutagenesis by PCR was utilized to introduce dinucleotide substitutions using the quick change sitedirected Mutagenesis Kit (Stratagene, La Jolla, CA). All constructs were sequenced as described previously to verify the fidelity of the mutagenesis.
Gel Mobility Shift Assays
Confluent Saos2 cells in T-75 flasks incubated for 3–24 h with PTH (10 nM) in α-MEM without serum were used to prepare nuclear extracts. The double-stranded oligonucleotides encompassing the inverted CCAAT (nts, −64 to −41, 5′-CGTGACAGTGATTGGCTGTTGGAA-3′), CRE1 (nts, −89 to −63, 5′-ATCCACGTTCTGACATCACCTTGGTCG), CRE2 (nts, −680 to −658, 5′-ATCAGCTGACCTCACATGCACGA-3′), AP1(1) (nts, −158 to −129, 5′-CGTTTCTTGTTTATTCAACTGAGCCTGTGT-3′), AP1(2) (nts, −493 to −469, 5′-CTTTAATTTATGAGTCAATGAAGTA -3′) and AP1(3) (nts, −809 to −785, 5′-TTGGTTTCATGAATCATTTCAAAAA-3′) in the human BSP promoter. For gel shift analysis the double-stranded oligonucleotides were end-labeled with [γ-32P] ATP and T4 polynucleotide kinase. Nuclear extract (3 µg) were incubated for 20 min at room temperature (20°C) with 0.1 pM radiolabeled double-stranded oligonucleotide in buffer containing 50 mM KCl, 0.5 mM EDTA, 10 mM Tris–HCl (pH 7.9), 1 mM dithiothreitol, 0.04% Nonidet P-40, 5% nondenaturing acrylamide gels (38:2 acrylamide/bis-acrylamide) run at 200 V at room temperature. After electrophoresis, the gels were dried, and the autoradiograms were prepared and analyzed using an image analyzer. Supershift experiments were performed using CRE binding protein 1 (CREB1 (CREB(1); sc-58), ATF4 (CREB(2); sc-200) (Santa Cruz Biotechnology, Inc.), CREB1 (CREB(2); p43, Rockland) and phospho-CREB1 (Ser133, Upstate)) antibodies. Anti-CREB1 antibodies react to CREB1 and phospho-CREB1. phospho-CREB1 antibody recognizes p43 phosphorylated CREB1. Antibody was added to the reaction mixture and incubated for 3 h at 4°C before electrophoresis was performed under the same conditions as described above.
Chromatin Immunoprecipitation (ChIP) Assays
Chip assays were carried out using a Chip-IT™ Express Enzymatic kit (Active Motif) according to the manufacturer's protocol. Briefly, Saos2 cells were grown to confluence in 100 mm dishes and cultured in α-MEM without serum for 12 h and incubated with or without PTH (10 nM) for 3 h. Saos2 cells were fixed for 10 min with 1% formaldehyde and then chromatin was prepared using the ChIP-IT Express Enzymatic Kit protocol. After washed by PBS, cell pellets were homogenized by dounce homogenizer and centrifuged to pellet the nuclei. The nuclei pellet was digested by the enzymatic shearing cocktail (200 U/ml) to shear the chromatin at 37°C for 5 min and the reaction was stopped with the addition of cold EDTA. The equivalent of 6.3 µg of DNA (sheared chromatin) was used as starting material (input) in each ChIP reaction with 2 µg of the appropriate antibody (CREB1 (p43, Rockland) and phospho-CREB1 (Ser133, Upstate)) and protein G magnetic beads at 4°C overnight. Place tube on magnetic stand to pellet beads on the tube side and wash the beads extensively. Elute chromatin from the beads by elution buffer and reverse cross link buffer, and then the samples were treated with proteinase K for 1 h at 37°C. The purified DNA was subjected to PCR amplification (1 cycle at 94°C; 5 min, and amplification was carried out for 30 cycles at 94°C; 30 s, 55°C; 30 s, 72°C; 30 s, and final extension at 72°C; 10 min) for the CRE1 and CRE2 site within the human BSP promoter using CRE1ChIP(1); 5′-CCTCTTGGCTCTAGAATCACGTTTC-3′, CRE1ChIP(2); 5′-CCCTCTCACTCACTCATTCACTTGC-3′, CRE2ChIP(3); 5′-CAATGGATCACTCCTGCTAGCTCTAG-3′, CRE2ChIP(4); 5′-CCACTGAGAGGCAGATTTTATATTTTG-3′ primers. The PCR products were separated on 2% agarose gels and visualized with ultraviolet light. All ChIP assays were repeated at least three times and with triplicate samples for each antibody used in ChIP reactions.
Statistical Analysis
Triplicate or quadruplicate samples were analyzed for each experiment, and experiments were replicated to ensure consistency of the responses to PTH. Significant differences between control and treatment were determined using unpaired Student's t-test.
RESULTS
Stimulation of BSP, Runx2 and Osterix mRNAs and BSP Protein Levels in Saos2 Cells
Saos2 cells were treated with PTH (10 nM) and changes in BSP mRNA and protein levels were analyzed by Northern and Western blots. Results of Northern blot showed that the PTH increased the BSP mRNA levels at 3 and 12 h (Fig. 1A). BSP protein levels were increased after stimulation by PTH (10 nM) at 3 and 6 h (Fig. 1B). Results of real-time PCR showed that the PTH (10 nM) increased the BSP (at 3 and 6 h) and Runx2 (at 6 h) mRNA levels (Fig. 2). Osterix mRNA expression did not increase statistically (Fig. 2).
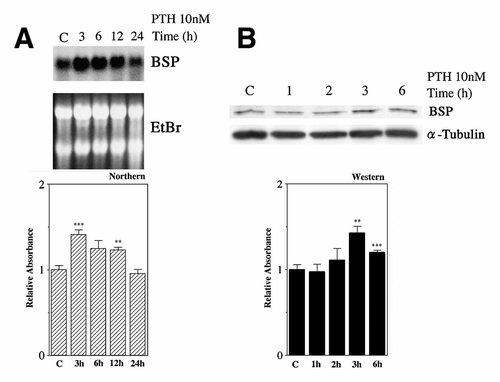
Effects of PTH on BSP mRNA and protein levels in Saos cells. A (upper panel): Saos2 cells were treated with or without PTH (10 nM) for 3, 6, 12, and 24 h. Then, total RNA was extracted, and the expressions of BSP mRNA in the cells were analyzed by Northern blot. The relative amounts of mRNA of BSP to 18S RNA (ethidium bromide staining) were calculated. The experiments were performed in triplicate for each data point. Results of a representative hybridization analysis for control and PTH treated cells are shown. B (upper panel): BSP protein expression in Saos2 cells. Cells were treated with or without PTH (10 nM) for 1, 2, 3, and 6 h, then cell lysate were prepared, and the expression of BSP was analyzed by Western blot using anti-BSP polyclonal antibody (LF-100). The experiments were performed in triplicate for each data point. A,B (Lower panel): Quantitative analyses of the triplicate data sets are shown with standard errors. Significant differences from control: **P < 0.05; ***P < 0.02.
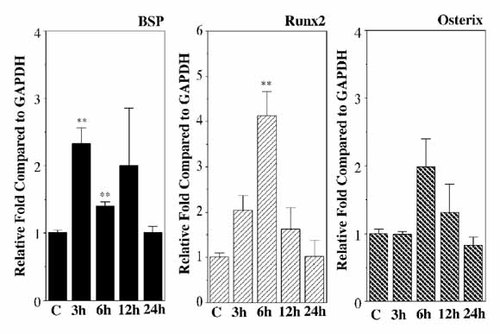
Effects of PTH on BSP, Runx2 and Osterix mRNA levels in Saos cells. The expressions of BSP, Runx2, Osterix and GAPDH mRNA in the Saos2 cells were measured by real-time PCR. The relative amounts of mRNA of BSP, Runx2 and Osterix to GAPDH were calculated. The experiments were performed in triplicate for each data point. Quantitative analyses of the triplicate data sets are shown with standard errors. Significant differences from control: **P < 0.05.
Transient Transfection Analyses of Human BSP Promoter Constructs
To determine the site of PTH-regulated transcription in the 5′-flanking region of the human BSP gene (Fig. 3), −184 to +60 (−184LUC), −211 to +60 (−211LUC), −428 to +60 (−428LUC), −868 to +60 (−868LUC) and −927 to +60 (−927LUC) human BSP promoter regions ligated to luciferase reporter genes were transiently transfected into Saos2 cells and the transcriptional activities determined in the presence or absence of PTH. PTH (10 nM) increased luciferase activities all of the constructs at 3 and 6 h (Fig. 4). Including within the DNA sequence that is unique in −184LUC to −927LUC, is an inverted CCAAT box (ATTGG; between −54 and −50), a cAMP response element 1 (CRE1; −79 to −72), an FGF2 response element (FRE; −96 to −89), an activating protein 1 response element (AP1(1); −148 to −142), a homeobox-binding site (HOX; −200 to −191) and cAMP response element 2 (CRE2; −674 to −667) in the human BSP gene promoter (Fig. 3). We previously identified a Pit-1 motif (−109 to −106) that mediated the stimulatory effects of PTH in the rat BSP gene promoter (Fig. 3; lower panel). However, there is no identical Pit-1 in the human BSP gene promoter (Fig. 3; a dotted line).
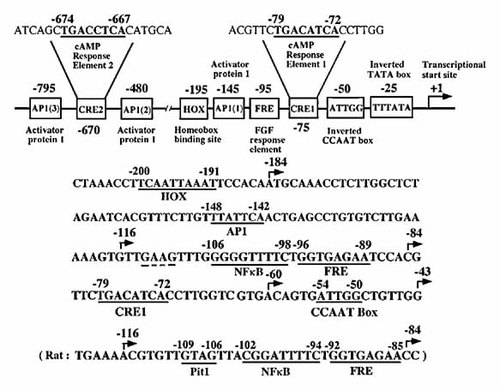
Regulatory elements in the proximal human BSP promoter. The position of the inverted TATA and CCAAT boxes, CRE1, FRE, HOX, CRE2 and three AP1 sites are shown in the proximal promoter region of the human BSP gene. The numbering of nucleotides is relative to the transcription start site (+1). The nucleotide sequences of two cAMP response element (CRE1 and CRE2) in the human BSP gene promoter are shown from −79 to −72 (TGACATCA) and −674 to −667 (TGACCTCA) (upper panel). The nucleotide sequences of the human BSP gene promoter encompassing an inverted CCAAT box, CRE1, FRE, NFκB, AP1 and HOX are shown from −208 to −43 (middle panel). The nucleotide sequence of rat BSP gene promoter (−121 to −83) encompassing FRE, NFκB and Pit-1 are shown (lower panel).
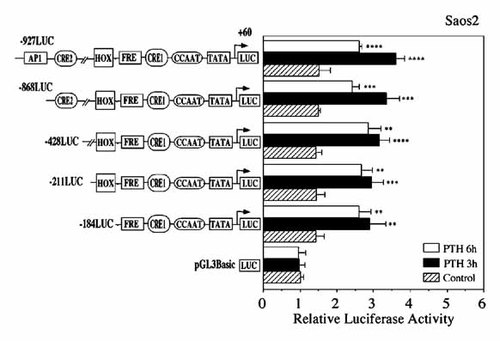
PTH up-regulates human BSP promoter activity in Saos2 cells. Transient transfections of Saos2 cells in the presence or absence of PTH (10 nM) for 3 and 6 h were used to determine transcriptional activity of chimeric constructs that included various regions of the human BSP promoter ligated to a luciferase reporter gene. The results of transcriptional activity obtained from four separate transfections with constructs, pGL3-basic and −184LUC to −927LUC, have been combined, and the values are expressed with standard errors. Significant differences from control: **P < 0.05; ***P < 0.02; ****P < 0.01.
Next, we introduced 2 bp mutations in CRE1 and CRE2 elements targeted by PTH within the −184LUC and −868LUC constructs. When mutation was made in the CRE1, PTH-induced −184LUC activity (−184mCRE1) was almost completely abolished, whereas PTH-induced −868LUC activity (−868mCRE1) partially inhibited (Fig. 5). Next we introduced mutation into the CRE2, PTH-induced luciferase activity in −868mCRE2 was partially abolished (Fig. 5). When mutations were made in the pairs of CRE1 and CRE2 in −868LUC (−868mCRE1/mCRE2), the effect of PTH on the luciferase activity was almost totally abrogated (Fig. 5). These results indicated that CRE1 and CRE2 act as functional response elements for PTH regulation of BSP gene transcription. PTH-induced BSP transcription (−184LUC and −868LUC) was inhibited by protein kinase A (PKA) inhibitor H89 (5 µM) and tyrosine kinase inhibitor herbimycin A (HA; 1 µM), and was not inhibited by protein kinase C inhibitor H7, indicating an involvement of PKA and tyrosine phosphorylation in the signaling pathway (Fig. 6).
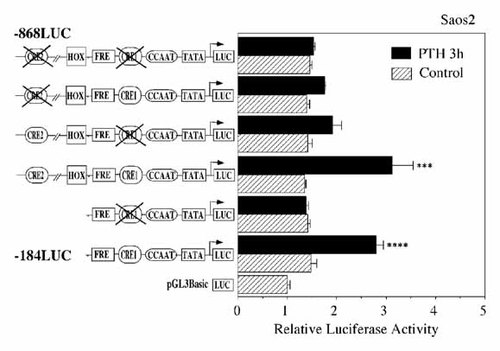
Site mutation analysis of luciferase activities. Dinucleotide substitutions were made within context of the homologous −184 to +60 (−184LUC) and −868 to +60 (−868LUC) BSP promoter fragment. mCRE1 (TGACAgaA), mCRE2 (TGACCgaA) and mCRE1/mCRE2 constructs were analyzed for relative promoter activity after transfection into Saos2 cells and examined for induction after treatment with PTH (10 nM) for 3 h. The results of transcriptional activity obtained from four separate transfections with constructs were combined and the values expressed with standard errors. Significant differences from control: ***P < 0.02; ****P < 0.01.
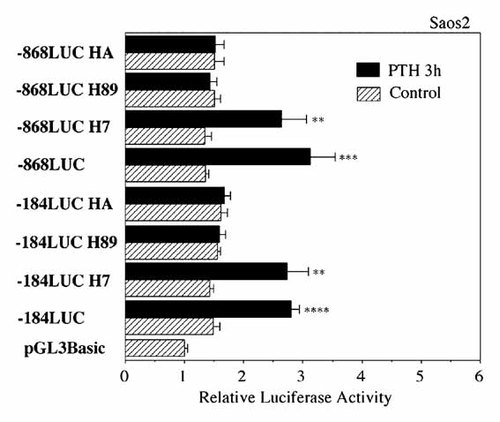
Effect of kinase inhibitors on transcriptional activation by PTH. Transient transfection analysis of −184LUC and −868LUC in the presence or absence of PTH (10 nM, 3 h) in Saos2 cells is shown together with the effects of the tyrosine kinase inhibitor (herbimycin A; HA, 1 µM), PKA inhibitor (H89, 5 µM) and PKC inhibitor (H7, 5 µM). The results of transcriptional activity obtained from four separate transfections with constructs were combined and the values expressed with standard errors. Significant differences from control: **P < 0.05; ***P < 0.02; ****P < 0.01.
Gel Mobility Shift Assays
To identify nuclear proteins that bind to the CRE1 and CRE2, double-stranded oligonucleotides were end-labeled and incubated with equal amounts (3 µg) of nuclear proteins extracted from confluent Saos2 cells that were either not treated (control) or treated with 10 nM PTH for 3, 6, and 12 h. When we used the inverted CCAAT sequence as a probe, the DNA–NF–Y protein complex [Shimizu and Ogata, 2002] did not change after PTH treatment (Fig. 7; lanes 1–4). With nuclear extracts from confluent control cultures of Saos2 cells, no shift of CRE1 and CRE2–protein complexes were evident (Fig. 7; lanes 5, 11). After stimulation by PTH (10 nM, 3, 6, and 12 h), CRE1–protein complex (single band) and CRE2–protein complexes (upper and lower bands) were increased (Fig. 7; lanes 6–8, 12–14). CRE1 and CRE2–protein complexes representing specific interactions were demonstrated by competition experiments in 40-fold molar excess of CRE1 and CRE2 reduced the DNA–protein complexes formation (Fig. 7; lanes 9 and 15). In contrast, mCRE1 and mCRE2 did not compete with CRE1 and CRE2–protein complexes formation (Fig. 7; lanes 10 and 16). To further characterize the proteins in the complexes formed with the CRE1 and CRE2, we used antibodies for several transcription factors. The CRE1–protein complexes showed single band (Fig. 8; lanes 2 and 3), was disrupted by two kind of CREB1 antibodies (Fig. 8; lanes 4 and 5) and supershifted by anti-phospho-CREB1 antibody (Fig. 8; lane 6). The CRE2–protein complexes showed CREB related doublet bands, and two bands below the doublet (Fig. 8; lanes 9 and 10). Upper band of the CREB related doublet was disrupted by two kind of CREB1 antibodies (Fig. 8; lanes 11 and 12) and supershifted by anti-phospho-CREB1 antibody (Fig. 8; lane 14). To verify that the PTH was operating through two CREs, we also used gel mobility shift analyses to evaluate the potential effects of PTH on the FRE, and three AP1 sites (Fig. 3). When we used the FRE and three AP1 elements (AP1(1), AP1(2) and AP1(3)) as probes, the FRE and three AP1–protein complexes did not change after PTH treatment (data not shown).
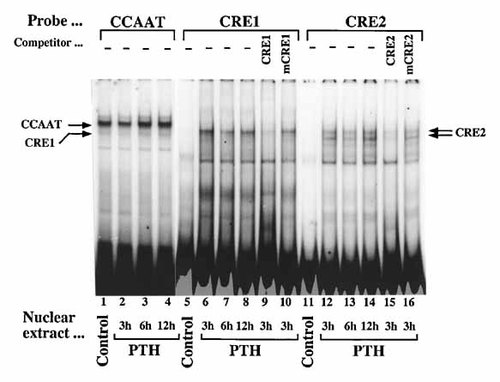
Gel mobility shift analysis of inverted CCAAT, CRE1, and CRE2 oligonucleotides. Radiolabeled double-stranded CCAAT (−64 CGTGACAGTGATTGGCTGTTGGAA −41), CRE1 (−89 ATCCACGTTCTGACATCACCTTGGTCG −63), CRE2 (−680 ATCAGCTGACCTCACATGCACGA −658) oligonucleotides were incubated with nuclear protein extracts (3 µg) obtained from Saos2 cells stimulated without (lanes 1, 5, and 11) or with PTH (10 nM) for 3 h (lanes 2, 6, 9, 10, 12, 15, and 16), 6 h (lanes 3, 7, and 13) and 12 h (lanes 4, 8, and 14). Competition reactions were performed using a 40-fold molar excess of unlabeled CRE1 (lane 9), mutation-CRE1 (mCRE1; −89 ATCCACGTTCTGACAgaACCTTGGTCG −63, lane 10), CRE2 (lane 15) and mutation-CRE2 (mCRE2; −680 ATCAGCTGACCgaACATGCACGA −658, lane 16) oligonucleotides. DNA–protein complexes were separated on 5% polyacrylamide gel in low-ionic-strength Tris-borate buffer, dried under vacuum and exposed to an imaging plate for quantitation using an image analyzer.
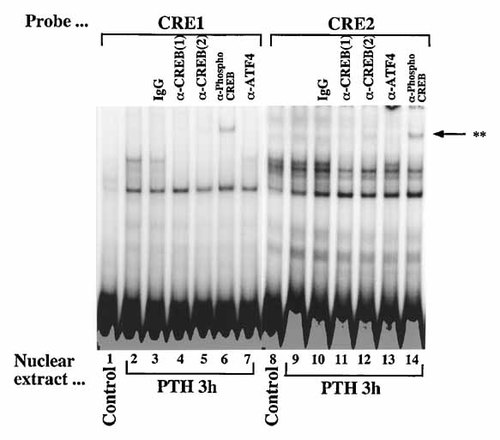
Specific binding of nuclear proteins to the CRE1 and CRE2. Radiolabeled double-stranded CRE1 and CRE2 were incubated with nuclear protein extracts (3 µg) obtained from Saos2 cells stimulated without (lanes 1 and 8) or with PTH (10 nM, lanes 2–7, 9–14) for 3 h. Supershift experiments were performed with 0.4 µg of antibodies against CREB(1), CREB(2), ATF4 and phospho-CREB added separately to each gel shift reaction.
ChIP Assay
We next examined whether CREB, phospho-CREB are able to interact directly with human BSP gene promoter and how PTH (10 nM) influences these transcription factors interaction with the CRE1 and CRE2. To clarify this issue further, we used ChIP assays to examine the in vivo association of these transcription factors with proximal CRE1 and distal CRE2 in Saos2 cells (Fig. 9). For this experiments, cells were treated with PTH (10 nM) for 3 h to induce BSP expression and cross-linked with formaldehyde. After enzymatic shearing, soluble chromatins were immunoprecipitated with either antibodies or control IgG. The PCR bands amplified with CRE1ChIP(1) and CRE1ChIP(2) and corresponding to DNA–protein complexes immunoprecipitated with antibodies revealed that CREB1 and phospho-CREB1 interacted with a chromatin fragment containing the CRE1 and phospho-CREB1 binding was increased after stimulation by PTH (Fig. 9, upper panel). When we used CRE2ChIP(3) and CRE2ChIP(4) for PCR amplification, CREB1 and phospho-CREB1 interacted with a chromatin fragment containing the CRE2 and phospho-CREB binding was also induced with PTH (Fig. 9, lower panel).
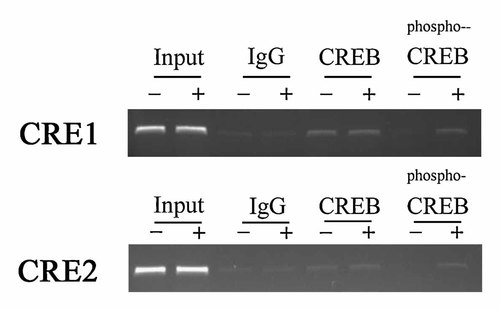
ChIP analysis of CREB1 and phospho-CREB1 binding to CRE1 and CRE2 sites in the human BSP promoter in Saos2 cells. Saos2 cells were grown to confluence and cultured in α-MEM without serum for 12 h and incubated without (−) or with (+) PTH (10 nM) for 3 h, before cells were cross-linked with formaldehyde for ChIP analysis. Three independent IP reactions were carried out with CREB1 and phospho-CREB1 antibodies and control reactions with normal rabbit IgG. Ethidium bromide stained agarose gels of the PCR products obtained with ChIP DNAs using the human BSP promoter primers CRE1ChIP(1) and CRE1ChIP(2) (top) and CRE2ChIP(3) and CRE2ChIP(4) (bottom). Input DNA was also used as control in PCR analysis.
DISCUSSION
In this study, we have identified that PTH response elements are two CREs (CRE1 and CRE2) in the human BSP gene promoter, which mediate PTH actions on BSP gene transcription. PTH stimulated BSP mRNA and protein expressions at 3 h (Fig. 1A,B). Using real-time PCR, BSP mRNA levels were increased at 3 h and Runx2 mRNA levels were induced at 6 h after stimulation by PTH (Fig. 2).
From transient transfection assays we initially located the PTH-responsive region to the proximal promoter (nt −184 to +60 (−184LUC); Fig. 4) of the BSP gene, which encompasses an inverted CCAAT box, a putative CRE, a FRE and an AP1 motif (Fig. 3). Transcriptional activity was totally abrogated when both CRE1 and CRE2 sites were mutated in −868LUC (Fig. 5). The involvement of the CRE1 and CRE2 elements is further supported by gel mobility shift analyses in which nuclear proteins formed complexes with the CRE1 and CRE2 elements that were increased by PTH (Fig. 7). There was no further increase in transcription in the −868LUC which contains a CRE2 site, compared with the −184LUC (Fig. 4), suggesting that the PTH activation of BSP transcription through the CRE2 might be weaker than that of on the CRE1. H89 and HA abrogated the transcriptional activities of −184LUC and −868LUC induced by PTH (Fig. 6).
PTH effect on osteoblasts is complex, involving the activation of multiple intracellular signaling pathways that can induce or suppress transcription of matrix proteins through different mechanisms. PTH increased osteocalcin gene expression through mRNA stabilization [Noda et al., 1988], suppressed osteopontin transcription without altering mRNA stability [Noda and Rodan, 1989]. PTH suppresses type I collagen gene expression in ROS 17/2.8 cells [Kream et al., 1989]. PTH stimulates cAMP levels in osteoblast-like cells [Majeska et al., 1980], and also increases PLC activity, inducing the formation of 1,4,5-IP3 and diacylglycerol. 1,4,5-IP3 triggers the release of Ca2+ from intracellular stores [Bidwell et al., 1991a,b], and diacylglycerol activates PKC [Berridge, 1993]. Consistent with this signaling pathway, the increased promoter activities (−184LUC and −868LUC) stimulated by PTH were abolished by the PKA inhibitor H89 (Fig. 6). On the other hand, PKC inhibitor H7 did not suppress PTH-induced BSP transcription. Tyrosine kinase mediated pathway appears to be involved (Fig. 6), since combinational effects of PKA and tyrosine kinase were crucial for BSP gene expression [Shimizu et al., 2006]. Further study is necessary to elucidate the signaling pathway after stimulation by PTH in human osteoblasts.
Previously, we reported a Pit-1 element in the rat BSP gene promoter was the target of PTH-stimulated transcription of the BSP gene [Ogata et al., 2000]. However, there is no identical Pit-1 sequence in the human BSP gene promoter (Fig. 3). PTH-induced transcriptional activity was totally abrogated when both CRE1 and CRE2 sites were mutated (Fig. 5). Therefore, two CREs in the human BSP gene promoter could be real response elements of PTH actions on human BSP gene transcription.
Transcription factors that bind specifically to CRE1 and CRE2 were demonstrated by the competition gel mobility shift, supershift and ChIP assays (Figs. 7-9). The results indicated that CRE1 and CRE2 sites physically interact with CREB1 and phospho-CREB1 in vitro by gel shift (Fig. 8) and in vivo using ChIP assays (Fig. 9). ChIP assays detected binding of CREB1 and phospho-CREB1 to a chromatin fragment containing CRE1 and CRE2 sites, and increased binding of phospho-CREB1 to the both sites. Most interestingly, a recent study by Huang et al. 2005 reported human osteocalcin and BSP expression is coordinated and regulated through cAMP-dependent PKA signaling and these promoter activities occurs through increased interaction between CRE and CRE-binding protein in human prostate cancer cells. Concerning about the Runx2 transcription factor, Barnes et al. [2003] demonstrated Runx2 is ectopically expressed and can activate BSP expression in human breast cancer cells. However, Runx2 and histone deacetylase 3 repress osteocalcin and BSP gene expression and that this repression is suspended upon osteoblastic cell differentiation [Schroeder et al., 2004; Lamour et al., 2007]. Results of Runx2 expression pattern in this study also indicated that Runx2 did not regulate BSP transcription directly.
In conclusion, our study has identified CRE1 and CRE2 sites in the human BSP gene promoter that mediate BSP transcription induced by PTH and that the PTH increased CREB1 and phospho-CREB1 bindings to the two CREs through PKA and tyrosine kinase pathways.
Acknowledgements
We thank Dr. Larry Fisher for kindly providing antibody. This work was supported in part by Grants-in-Aid for Scientific Research (B; No. 18390563) from the Japan Society for the Promotion of Science (JSPS), a Grant for Supporting Project for Strategic Research by the Ministry of Education, Culture, Sports, Science, and Technology, 2008-2012.




