Cloning and analysis of rat osteoclast inhibitory lectin gene promoter†
Jin-Xing Quan and Fang Zheng contributed equally to this work.
Abstract
Osteoclast inhibitory lectin (OCIL) is a novel regulator of bone remodeling, however, little is known concerning how OCIL is regulated to date. In this study, approximately 4.4 kb of the 5′-flanking sequence of rat OCIL gene was cloned into the promoter-less reporter vector pGL3-basic and transiently transfected into three different cell lines. The differences in the levels of luciferase activity paralleled well with the levels of OCIL mRNA expression in these cells, suggesting that the regulation of rat OCIL gene expression occurs mainly at the transcriptional level. Additional luciferase assays using a series of constructs containing unidirectionally deleted fragments showed that the construct-1819/pGL3 (−1819 to +118) exhibited the highest luciferase activity, suggesting the presence of functional promoter in this region. The region from −4370 to −2805 might contain negative regulatory elements, while the region from −1819 to −1336 might have important positive regulatory elements that enhance OCIL transcription. Sequence analysis of the promoter revealed the absence of both TATA and CAAT boxes. However, in the proximal promoter region (−81 to +118), several potential transcription factor binding sites that may be responsible for the basal transcriptional activity of rat OCIL promoter were observed. The promoter contains several potential Sp1 binding sites, and cotransfection of a shRNA expression plasmid that knockdowns Sp1 significantly reduced OCIL promoter activity and endogenous gene expression and moreover, overexpressing Sp7, a Sp1 family member that also binds to Sp1 binding sequence, increased OCIL promoter activity and gene expression, suggesting a role of Sp1 family proteins in regulation of OCIL transcription. J. Cell. Biochem. 106: 599–607, 2009. © 2009 Wiley-Liss, Inc.
Bone remodeling results from the coordinate activities of osteoblasts and osteoclasts. An imbalance in the bone remodeling process results in metabolic bone diseases such as osteoporosis or osteopetrosis [Suda et al., 1997]. The osteoblasts and osteoclasts are regulated by different hormones and by locally produced growth factors, cytokines, and prostaglandins. Also, the two cell types regulate each other by cell–cell interaction mediated by receptor activator of NF-κB ligand (RANKL) on the osteoblasts and receptor activator of NF-κB (RANK) on the osteoclast surface [Lacey et al., 1998; Nakagawa et al., 1998]. Osteoprotegerin (OPG), the decoy receptor of RANKL, is critical for its ability to bind RANKL and inhibit osteoclast formation [Lacey et al., 1998]. Up to date, the essential role of OPG/RANKL/RANK system in the process of osteoclast maturation as well as activation has been well established, and the majority of other bone resorption regulators regulate osteoclast formation and activation through their effects on this system.
Recently, some protein inhibitors of osteoclast formation have been discovered that might act independently of the RANKL/RANK signaling pathway [Horwood et al., 1998, 2001; Choi et al., 1999]. Among these is osteoclast inhibitory lectin (OCIL). Up to date, the OCIL cDNA of human, mouse and rat have been identified [Zhou et al., 2001, 2002; Hu et al., 2004], and structurally they belong to type II transmembrane molecules with a C-type lectin extracellular domain. Similar to RANKL, OCIL is highly expressed in osteoblasts, chondrocytes, and other extraskeletal tissues such as kidney, liver, gut, heart, and skeletal muscle. OCIL dose-dependently inhibited multinucleate osteoclast formation from adherent murine spleen cells treated with RANKL and M-CSF. Such an effect relies on the predominant action of OCIL to inhibit the proliferation of mononuclear osteoclast precursors [Zhou et al., 2001, 2002]. These evidences strongly suggested that OCIL might have an action to oppose RANKL in the control of osteoclastogenesis. Furthermore, the latest data showed that OCIL inhibits osteoblast differentiation and function in vitro [Nakamura et al., 2007], highlighting a complicated but crucial role, played by OCIL in bone remodeling process. A variety of osteotropic factors in osteoblastic cells, including retinoic acid, 1,25-dihydroxyvitamin D3, prostaglandin E2, parathyroid hormone, parathyroid hormone-related protein, interleukin-1α, and interleukin-11, upregulate OCIL mRNA expression in osteoblast-like cells [Zhou et al., 2001; Zheng et al., 2009].
The way OCIL is regulated by these factors should profoundly influence bone remodeling. However, little information is available to date concerning how the expression of this gene is controlled in osteoblast-like cells. This is mainly due to the lack of information on transcriptional regulation of the OCIL gene. Thus, characterization of promoter sequences and identification of cis-elements and trans-acting factors that regulate the OCIL gene transcription may greatly contribute to understanding of molecular mechanisms of OCIL regulation. As a first step toward the better understanding of molecular mechanisms of the OCIL gene expression, we cloned the 5′-flanking sequence of rat OCIL gene and identified the regulatory region essential for transcriptional activation in osteoblast-like cells.
MATERIALS AND METHODS
Cell Culture and Reverse Transcription-Polymerase Chain Reaction
Rat UMR106 osteoblast-like cells and rat C6 glioma cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Hyclone) supplemented with 10% fetal calf serum (GIBCOL BRL), 100 µg/ml penicillin, and 100 U/ml of streptomycin. Both cultures were maintained at 37°C in a humidified 5% CO2-containing atmosphere.
Primary osteoblastic cell cultures were prepared from calvaria of day 4 neonatal Wistar rats. Briefly, the calvaria were digested with a mixture of trypsin and collagenase for five 10-min intervals. The cells from digestions 3–5 were harvested and cultured in T-25 flasks in complete αMEM medium containing 10% fetal calf serum, L-ascorbate (100 µg/ml), and antibiotics.
Total RNA was extracted from the cell cultures with TRIZOL (Invitrogen) and then converted to cDNA using M-MLV Reverse Transcriptase (Promega). PCR reactions were performed for 32 (OCIL) or 25 (β-actin) cycles of denaturation at 94°C for 30 s, primer annealing at 58°C for 30 s, and extension at 72°C for 45 s. The primers for OCIL were 5′-CTGAGTATAACAACTCGGTTTC-3′ (sense) and 5′-GTGTTCCTCATGACTGTTAG-3′ (antisense), yielding a 190 bp fragment. The primers for the internal control gene β-actin were 5′-GAACCCTAAGGCCAACCGTG-3′ (sense) and 5′-AGGCATACAGGGACAACACAGC-3′ (antisense), yielding a 104 bp fragment. The amplified products were run on a 1.5% agarose gel.
Rat Genome DNA Isolation, Computer Assistant Analysis, and Cloning of 5′-Flanking Region of OCIL Gene
Genome DNA was isolated from the brain tissue of a Wistar rat using a DNA extraction kit (TaKaRa), and was dissolved in water.
The full-length rat OCIL cDNA sequence (accession number: AF 321552) was identified by Zhou et al. 2001, and in that study the 5′ ends of the cDNA and the transcription initiation site were determined by 5′ RACE strategy. Based on the DNA sequence (ID 113937) of rat OCIL, a pair of specific PCR primers were designed to amplify a 4488 bp DNA fragment containing 118 bp of exon 1 and 4370 bp of 5′-flanking region of rat OCIL gene. The primers used are 5′-TAGTACTCCTAACACTTCTTGGGACTG-3′ (sense) and 5′-AAGCTTGACAGACCACAGCTGCTAC-3′ (antisense). The underlined HindIII sequence is to be used for cloning. The PCR amplification was run using the high-fidelity LA Taq DNA polymerase (TaKaRa) in a Perkin Elmer 9600 thermocycler programmed for an initial denaturation step of 3 min at 95°C followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 3 min. The PCR product was gel purified and subsequently cloned using PCR cloning vector pTA2 (TOYOBO, Japan), and the resulting recombinant plasmid, termed −4370/pTA2, was sequenced to confirm the presence of the desired sequence.
Homology searches were performed using BLAST (Basic Local Alignment Search Tool) from the National Centre for Biotechnology Information at http://www.ncbi.nlm.nih.gov. The promoter sequence was analyzed by using the Transcription Element Search System (TESS of the Computational Biology and Informatics Laboratory, School of Medicine, http://agave.humgen.upenn.edu/tess/index.html) with a 6-bp minimum element size limit, a 5% mismatch allowance, a minimum log-likelihood of homology of 10, and a secondary log-likelihood density threshold of 1.6. Further analysis and prediction of transcription factor binding sites was performed with TFSEARCH (http://pdap1.trc.rwcp.or.jp/research/db/TFSEARCH.html).
Establishment of Luciferase Reporter Gene Constructs
Since there exists an EcoRV recognition sequence within the multiple cloning sites of pTA2 vector, the recombinant plasmid −4370/pTA2 was digested using EcoRV and HindIII. The released fragment was cloned into the promoter- and enhancer-less luciferase reporter vector pGL3-basic (Promega) at the sites of SmaI and HindIII, and thus located upstream of the firefly luciferase coding sequence. Then, using −4370/pTA2 as the template, various lengths of DNA fragments upstream of the initiating ATG codon were PCR amplified in the presence of high-fidelity Primestar DNA polymerase (TaKaRa). The primers used are summarized in Table I. It is noteworthy that the common antisense primer contains a HindIII site at the 5′ end. The amplified fragments were digested with HindIII enzyme, and inserted into pGL3-basic at SmaI and HindIII sites. The name of each reporter construct was assigned according to the 5′-end nucleotide number of the inserted promoter sequence, upstream or downstream of transcription start site.
| Name of fragment | DNA sequence (from 5′ to 3′) | Position |
|---|---|---|
| Rocil-2805 | GATAACAATACCAGGCTGTA | −2805 to −2786 |
| Rocil-1819 | CGACTTGCCAGTTCTCAT | −1819 to −1811 |
| Rocil-1336 | GGGGTAGGAATGTATTTTTCT | −1336 to −1316 |
| Rocil-721 | ATTCTCCCTCTTCCTTAGGA | −721 to −702 |
| Rocil-400 | TTCCTGTATGTCTGTGTGC | −400 to −382 |
| Rocil-168 | TTCCCAATTATTGCTAG | −168 to −152 |
| Rocil-81 | ATGGTTGGAAACACTGTT | −81 to −64 |
| Rocil-22 | CATGTCTCCCTTGAATCCT | −22 to −3 |
| Rocil-+37 | ATCCTCCCCCACTCCTCTCC | +37 to +56 |
| Antisense primer | AAGCTTGACAGACCACAGCTGCTAC | +99 to +118 |
- The underlined letters represent the HindIII site.
Transient Transfections and Luciferase Assays
Exponentially growing cells were used for transfection assays. Twenty-four hours after plating on 24-well plates, the cells at 50–70% confluence were transfected using lipofectAMINE 2000 reagent. For each transfection, 0.8 µg of each promoter construct and 2 µl lipofectAMINE 2000 reagent were used. 0.1 µg of the pSV-β-galactosidase (Promega) vector was cotransfected into cells to normalize the variations in transfection efficiency. The promoter- and enhancer-less pGL3-basic vector was used as negative control. Cells were harvested 48 h after transfection and luciferase activity was determined using the Luciferase Assay System (Promega). Meanwhile, β-galactosidase activity was assayed by using β-Galactosidase Enzyme Assay System (Promega). We normalized variations in transfection efficiency by dividing the measurement of the luciferase activity by that of the β-galactosidase activity. The relative luciferase activity was then calculated by dividing the normalized luciferase activity by that obtained with the parent pGL3-basic vector. All of the data shown in this study were obtained from at least three independent experiments.
RNA Interference
shRNA construct that targets Sp1 gene was constructed using a previously described protocol [Li et al., 2008]. The insert sequence was as follows: 5′-GATCCGTGAGAACAGCAACAACTCCTTCAAGAGAGGAGTTGTTGCTGTTCTCATTTTTTGGAAA-3′, where the underlined part indicates the sense strand of the target sequence. The insert was cloned into the shRNA expression vector pSilencer 3.1-H1 neo at the sites of BamHI and HindIII. To examine whether the construct knockdowns Sp1 and affects OCIL expression, UMR106 was transfected with 0.8 µg of the construct, or the negative control vector. Forty-eight hours after the transfection, total RNA was isolated from these cells and reverse transcribed. Sp1 mRNA level was examined to confirm the correct knockdown of Sp1 using PCR that was performed for 30 cycles of denaturation at 94°C for 30 s, primer annealing at 56°C for 30 s, and extension at 72°C for 30 s. The primers used were 5′-GAATGCTGCTCAACTGTCCTC-3′ (sense) and 5′-TCTCCACCTGCTGTCTCATC-3′ (antisense). OCIL mRNA level was examined by PCR using above described protocol. To examine whether the construct affects the OCIL promoter activity, 0.6 µg of this construct or the negative control vector pSilencer 3.1-H1 neo, was cotransfected with −1819/pGL3 (0.6 µg/well) into UMR106 cells. Cells were harvested 48 h after transfection and luciferase activity was examined and normalized to the amount of protein in the cell layer.
Gene Overexpression
The expression construct harboring the full-length Sp7 open reading frame and a Kozak sequence before the initiating methionine was established previously in our lab by Dr. Peng-Cheng Xu (Division of Nephrology, Tianjin Medical University Hospital). To examine whether the construct overexpresses Sp7 (named pcDNA3/Sp7) and affects OCIL expression, UMR106 was transfected with 0.8 µg of the construct, or the empty vector pcDNA3. The cells stably expressing the plasmids were used for RT-PCR analysis of Sp7 and OCIL expression. The PCR amplification for Sp7 was performed for 32 cycles of denaturation at 94°C for 30 s, primer annealing at 56°C for 30 s, and extension at 72°C for 45 s using the primers 5′-ATGGCGTCCTCTCTGCTTG-3′ (sense) and 5′-TACCAGGAGCCGTAGGGATG-3′ (antisense). To examine whether the construct affects the OCIL promoter activity, 0.6 µg of this construct or the empty vector pcDNA3, was cotransfected with −1819/pGL3 (0.6 µg/well) into UMR106 cells. Cells were harvested 48 h after transfection and luciferase activity was determined.
Statistical Analysis
Statistical analysis was performed by “t” test or one-way analysis of variance (ANOVA) and Student–Newman–Keuls test when ANOVA demonstrated significances. A P-value of <0.05 was regarded as significant.
RESULTS
Cloning and Computer Analysis of Rat OCIL Promoter
The complete cDNA sequence of rat OCIL gene was identified by Zhou et al. 2001, and in their study, the transcriptional initiation site for rat OCIL was mapped with 5′ RACE strategy. Their results demonstrated that the full-length cDNA sequence of rat OCIL gene is 1628 bp, which encodes a protein of 233 amino acids, and the translation start codon is at the position of nt +186 (Fig. 1).

Structural analysis of the rat OCIL 5′-flanking region. Potential binding sites for transcription factors are underlined and designated. The transcription start site is indicated by the first letter in the sequence in bold and black. The ATG codon that initiates translation is at the end of the sequence. The numbers of nucleotides upstream of transcription start site (+1) are shown on the left of the sequence.
In the present study, we wished to clone and characterize the OCIL gene promoter. Genome DNA was isolated from rat brain tissues using standard methods. And then, we successfully amplified a 4488 bp DNA fragment containing 118 bp of exon 1 and 4370 bp of the 5′ flanking region of rat OCIL. Sequence analysis of the cloned PCR product revealed that this sequence had complete homology with the expected 5′-flanking DNA sequence of rat OCIL gene.
We then analyzed the DNA sequence using the TFSEARCH and Transcription Element Search System (TESS). The results showed that the proximal 5′-flanking region of rat OCIL gene is relatively GC-rich (54% in the region from −81 to +37) and lacks basal elements such as a TATA box, a CAAT box, or an initiator sequence [Javahery et al., 1994]. However, a number of potential transcription factor binding sites proximal to the transcription start site were predicted, including Sp1 at position −37 and −60, MZF1 at position −41, GATA-1 at position −32, and NF-AT at position −78. In addition, an AP1, AP4, and an AREB6 potential binding sites were observed immediately downstream of the transcription start site (Fig. 1).
Construction of Plasmids and Functional Characterization of the 5′-Flanking Region
To examine the transcriptional activity of the rat OCIL gene promoter, we first inserted the 4488-bp fragment (−4370 to +118) upstream of the firefly luciferase gene in the pGL3-basic vector. The resultant plasmid −4370/pGL3 was transiently transfected into three endogenous OCIL mRNA-expressing cells, that is, UMR106 osteoblast-like cells, primary calvarial osteoblastic cells, and C6 glioma cells. The firefly luciferase activity obtained was normalized to the β-galactosidase activity and compared with that from the pGL3-basic vector. The −4370/pGL3 showed significant highest activity in UMR106 cells, that is, 32-fold greater than the baseline activity of the pGL3-basic vector. In primary osteoblastic cells and C6 cells, the vector −4370/pGL3 exhibited significant activity as well, that is, 16- and 18-fold, respectively greater than pGL3-basic vector (Fig. 2). The differences of luciferase activity were comparable with the levels of OCIL mRNA in the three cells as shown in Figure 3, where UMR106 exhibited a higher mRNA level than the other two cells, making our results convincing.
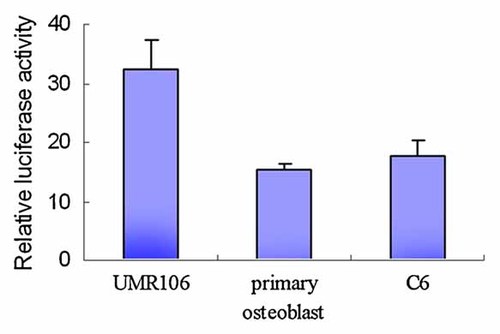
Relative luciferase activity of the −4370/pGL3 construct. The luciferase reporter −4370/pGL3 was transiently transfected into rat UMR106 osteoblast like cells, primary osteoblastic cells, or C6 glioma cells. The pGL3-basic plasmid was used as negative control. Cells were harvested 48 h after transfection and luciferase activity was measured. Relative luciferase activity of the construct was calculated as described in Materials and Methods Section. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
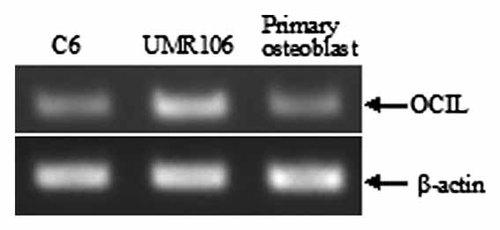
OCIL mRNA expression levels in three different cells. OCIL mRNA expression in UMR106 osteoblast-like cells, primary calvarial osteoblastic cells, and C6 glioma cells was analyzed by using RT-PCR. The photograph of the PCR analysis from a representative experiment is shown.
To further determine the elements responsible for the rat OCIL promoter, we next made a series of luciferase constructs containing unidirectionally deleted fragments from the −4370/pGL3 (Fig. 4). All deletion derivatives were obtained by PCR amplification of 5′ deletion fragments of OCIL followed by inserting the PCR products into the pGL3-basic vector. The various constructs were transiently transfected into UMR106 cells and luciferase activity was measured. The results showed that the deletion from nt −4370 to −2805 (−2805/pGL3) markedly increased the luciferase activity to 153-fold higher than that of the pGL3-basic. Further deletion to nt −1819 (−1819/pGL3) showed the highest activity, which was approximately 184-fold higher than the activity of the pGL3-basic, suggesting that the core promoter is present in this region (−1819 to +118), and that the sequence between −4370 and −2805 might contain negative elements that suppress the promoter activity. However, further deletion to nt −1336 (−1336/pGL3) significantly reduced the promoter activity to only 5.3-fold greater than that of pGL3-basic, suggesting the presence of major positive elements in the region (−1819 to −1336) that enhance the basal promoter activity. Among the shortest constructs, −721/pGL3 had an 11.9-fold higher activity, −400/pGL3 (−400 to +118) a 6.1-fold, −168/pGL3 (−168 to +118) an 11.2-fold, and −81/pGL3 (−81 to 118) an 11.8-fold higher activity than pGL3-basic, while −22/pGL3 (−22 to +118) and +37/pGL3 (+37 to +118) produced similar activity to that of pGL3-basic vector. These data suggest that the sequence between −81 and −22 might contain regulatory elements that contribute to the basal transcriptional activity of the OCIL gene, thus the region from −81 to +118 could be referred as the proximal promoter.
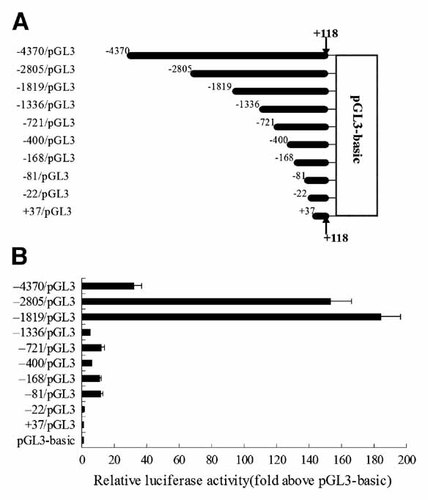
Deletion analysis of the rat OCIL promoter. A: Schematic view of rat OCIL promoter luciferase reporter constructs; black columns represent the position and extent of genomic DNA attached to the pGL3-basic reporter gene. All 5′ deletion constructs were named as shown on the left. B: Various 5′ deletion constructs were transiently transfected into rat UMR106 cells, and cells were harvested 48 h after transfection and luciferase activity was measured.
Sp1 and Sp7 Activate the OCIL Promoter and Induce Endogenous OCIL Expression in UMR106 Cells
Sp1 shRNA construct and Sp7 overexpression construct were established and confirmed to be capable of functioning properly in the UMR106 cells. While Sp1 shRNA construct dramatically reduced Sp1 level, Sp7 overexpression construct potently increased Sp7 level (data not shown). To investigate whether Sp1 family members can transactivate the OCIL promoter, Sp1 shRNA construct and Sp7 overexpression construct were cotransfected, respectively with the −1819/pGL3 promoter reporter construct into UMR106 cells. Knockdown of Sp1 potently reduced −1819/pGL3 promoter activity by about 40%, as compared to the negative control transfected cells (Fig. 5). On the other hand, Sp7 overexpression induced −1819/pGL3 promoter activity by 1.9-fold, as compared to the empty vector pcDNA3 transfected cells (Fig. 6). Furthermore, we found that Sp1 knockdown significantly reduced the endogenous mRNA levels of OCIL (by about 50%) (Fig. 5), whereas, Sp7 overexpression significantly increased the endogenous mRNA levels of OCIL in UMR106 cells (by about twofold) (Fig. 6). These results indicate that Sp1 and Sp7 transcription factors might upregulate the OCIL gene through Sp1 binding sites in the promoter region.
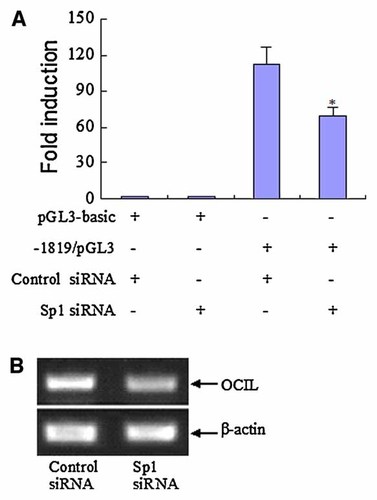
Sp1 regulation of rat OCIL transcription. A: Sp1 shRNA construct, or the control siRNA vector pSilencer 3.1-H1 neo was cotransfected with the reporter plasmid −1900/pGL3, or the parent vector pGL3-basic, and luciferase assays were performed using UMR106 cells. Luciferase activity relative to that of the control siRNA plus pGL3-basic transfection was described as fold induction of transcription. * Significant versus control siRNA plus −1900/pGL3 transfection, P < 0.05. B: UMR106 was transfected with Sp1 shRNA construct, or the negative control vector. OCIL mRNA expression was analyzed by using RT-PCR. The photograph of the PCR analysis from a representative experiment is shown. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
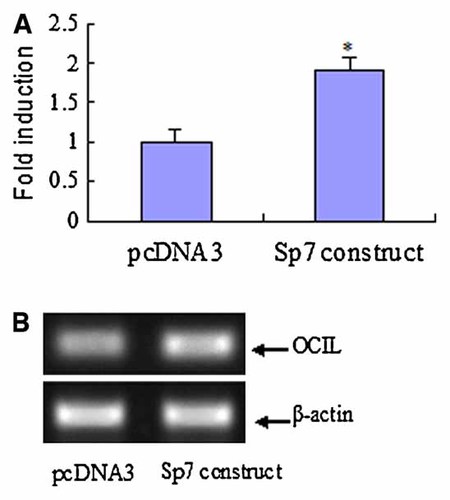
Sp7 regulation of rat OCIL transcription. A: Sp7 overexpression construct was cotransfected with the reporter plasmid −1900/pGL3, and luciferase assays were performed using UMR106 cells. As effector control, the empty plasmid pcDNA3 was used. Luciferase activity relative to that of the control was described as fold induction of transcription. * Significant versus pcDNA3 transfection, P < 0.05. B: UMR106 was transfected with Sp7 overexpression vector, or the empty vector pcDNA3. OCIL mRNA expression was analyzed by using RT-PCR. The photograph of the PCR analysis from a representative experiment is shown. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
Despite the important role OCIL plays in the process of bone remodeling, little is known about the transcriptional regulation of OCIL gene. Investigation of the molecular mechanisms underlying OCIL gene expression could lead to a better understanding of OCIL function. The cloning and characterization of the OCIL gene promoter in this study is an essential step toward this goal. We isolated and identified 4370 bp of the 5′-flanking region of the rat OCIL gene (Fig. 1). Our luciferase assays using three different cells that differ in the levels of OCIL expression revealed that this cloned 5′-flanking sequence had definite transcriptional activity, and the differences of the activity in these three cells paralleled quite well with their mRNA expression levels. The data also suggest that the regulation of OCIL gene expression occurs mainly at the transcriptional level. Thus, identification of the cis-elements that regulate the OCIL gene transcription is important for understanding the regulation of this gene.
In general, the core promoters of mammalian protein-coding genes often contain a TATA box, located 25–30 bp upstream of the transcription start site, and/or an initiator (Inr) element, which overlaps the transcription start sites of many TATA-containing and TATA-lacking promoters. Each element can independently direct basal transcription by RNA polymerase II and can determine the location from which transcription initiates [Javahery et al., 1994; Lo and Smale, 1996; Smale, 1997].
TFSEARCH and TESS analysis of the 5′-flanking sequence revealed that rat OCIL promoter lacks a canonical TATA box, a CAAT box, and the consensus sequence (PyPy A + 1 N T/A PyPy) for an initiator [Lo and Smale, 1996], suggesting that rat OCIL promoter belongs to the class of TATA- and Inr-less promoter. Originally, TATA-less promoters were thought to be common to constitutively active genes and expressed at a relatively constant level in all tissues, in an unregulated fashion. However, recent studies showed that vast majority (76%) of human core promoters lack TATA-like sequences and 46% of human and yeast promoters lack both TATA and Inr elements [Bajic et al., 2004; Yang et al., 2007]. These results suggest that additional core promoter motifs may play a key role in specific transcription initiation at a significant number of human and yeast promoters that lack both TATA and Inr elements. There are strong evidences demonstrating that the transcription of TATA- and Inr-less promoters can be activated either by transcription factor Sp1 in most cases [Hasan and MacDonald, 2002; Kim et al., 2002; Andersen et al., 2004; Le Mée et al., 2005; Zeng et al., 2005; Dong et al., 2008], or in fewer cases, by several other transcription factors that do not belong to Sp family members [Miyamoto et al., 1998; Kam et al., 2005]. It has long been known that Sp1-binding sites (i.e., GC-boxes) are frequently found in TATA-less promoters within CpG islands and that Sp1 can be located on the sense and antisense strands [Mieda et al., 1996], which can direct transcription initiation from core promoters lacking both TATA and Inr elements [Smale and Kadonaga, 2003].
The overall GC content is about 54% and there are no CpG islands present in the proximal region of the OCIL promoter. However, a GA-rich (GA/CT box) region (from −61 to −29), which may potentially bind a transcription factor of the zinc finger family (i.e., Sp1 and MZF-1), is located closely upstream of the transcription start site, and two potential binding sites for Sp1, one site for myeloid zinc finger-1 (MZF-1) were observed in this region (Fig. 1). Of particular note is the consensus sequence (GGGAGGG from −37 to −31) of Sp1, which overlaps the MZF-1 site (GAAGGGGA from −41 to −34), has a minor difference from the canonical sequence (GGGCGGG) of Sp1. However, it has been confirmed to have the high affinity to Sp1 proteins and is responsible for the basal transcription activation of human mitochondrial glycerol phosphate dehydrogenase promoter B and rat immune-associated nucleotide 4-like 1 promoter [Andersen et al., 2004; Le Mée et al., 2005]. MZF-1 belongs to the zinc finger protein family, and has been known to play a crucial role in either the basal transcription activation or enhancement of transcription of several genes [Morris et al., 1994; Andersen et al., 2004; Le Mée et al., 2005; Zeng et al., 2005]. Besides this, one site for transcription factor NF-AT was also found in this region, which was previously found in the promoter region of several genes and identified to be responsible for the enhancement of transcriptional activity of such a TATA-less promoter [Seth et al., 2003; Le Mée et al., 2005].
Through luciferase reporter assays, we found that the luciferase reporter constructs harboring various lengths of OCIL 5′-flanking sequence displayed different levels of luciferase activity, and the highest luciferase activity of rat OCIL promoter deletions was approximately 184-fold greater than that of the promoter-less pGL3-basic vector, indicating the presence of a functional and strong promoter in the cloned 5′ flanking fragment of OCIL. Among the shorter deletion constructs, −81/pGL3 (−81 to +118) had an 11.8-fold higher activity than pGL3-basic, while −22/pGL3 (−22 to +118) and +37/pGL3 (+37 to +118) had no promoter activity, suggesting that the region between nt −81 and −22 very likely contains important cis-acting elements that are responsible for the basal activity of rat OCIL promoter. These results were strongly supported by the finding that the region from nt −81 to −29 contain two Sp1 sites, one MZF-1 site and one NF-AT site, all of which were previously identified to be responsible for the basal as well as enhanced promoter activity of lots of genes. Thus, the region from −81 to +118 could be referred as the proximal promoter and these sites, especially the Sp1 and MZF-1, may contribute to the basal promoter activity of rat OCIL.
The highest activity was observed with the −1819/pGL3 (−1819 to +118), which exhibited 184-fold higher luciferase activity as compared with that of pGL3-basic, while the full length luciferase construct, −4370/pGL3 (−4370 to +118), had 32-fold promoter activity in UMR106 cells. However, deletion from −4370 to −2805 (−2805/pGL3) restored the promoter activity to 153-fold greater than that of pGL3-basic vector, suggesting the presence of negative regulatory elements in this region. Consistent with this hypothesis, in the region from −4370 to −2805, computer analysis with TESS software indicated the presence of three delta EF1 repressor sequences at position −4077, −3379, and −3188 [Sekido et al., 1994]. Further deletions from −1819 to −1336 (−1450/pGL3) resulted in significant loss of promoter activity, suggesting presence of important positive regulatory elements in this region that enhance rat OCIL transcription. Computer analysis revealed that this region contains the highest density of possible cis-acting elements, including two Sp1 site, one C/EBP site, one AP4 site, two GATA-1 sites, one Oct-1 site, two AREB6 sites, one NF-Y site, and one NF-AT site (Fig. 1).
Since there exist several potential Sp1 binding sites in the OCIL promoter, we next studied whether Sp1 family transcription factors can transactivate the promoter and regulate OCIL expression. It has been reported that several Sp1 family proteins, including Sp1 and Sp7/osterix, are expressed in bone [Nakashima et al., 2002; Liu et al., 2005]. Sp1 shows ubiquitous expression patterns and regulates a variety of cellular activities including cell growth, differentiation, apoptosis, and oncogenesis [Black et al., 2001]. However, the significance of Sp1 in bone physiology and pathology remains to be clarified. In contrast, Sp7/osterix is a bone-specific transcription factor essential for osteoblast differentiation and bone formation [Nakashima et al., 2002; Koga et al., 2005; Matsubara et al., 2008]. Like other Sp1 family proteins, Sp7 also binds to G/C-rich Sp1 binding sequences although its amino acid sequence shares no significant similarity with other Sp1 family members. In the present study, functional analysis by luciferase constructs and RT-PCR showed that overexpressing Sp7 in UMR106 significantly increased OCIL promoter activity and endogenous OCIL mRNA levels (through functional Sp1 binding sites). Meanwhile, knockdown of Sp1 by siRNA decreased OCIL promoter activity and endogenous gene expression. Taken together, we conclude that Sp1 family proteins are involved in the transcriptional regulation of the rat OCIL gene in the osteoblast-like cells.
In summary, we report here for the first time the cloning and characterization of the rat OCIL promoter. The 5′-flanking region of rat OCIL gene was cloned in which we have confirmed the presence of a functional promoter. Through luciferase reporter assays and computer analysis, the proximal promoter of rat OCIL and several regions that might contain the important positive or negative regulatory elements and a variety of potential transcription factor binding sites have been identified. Sp1 family proteins such as Sp1 and Sp7 have an important role in the regulation of rat OCIL promoter activity.
Acknowledgements
This work was supported by a project of Natural Science Foundation of China (No. 30300171), a key project of Tianjin Municipal Natural Science Foundation (No. 07JCZDJC07500) and also a key Project of Chinese Ministry of Education (No. 208011).




