Gene expression profiling of macrophages following mice treatment with an immunomodulator medication
Abstract
Canova (CA) is a complex homeopathic medication used in diseases where the immune system is depressed. Previous studies demonstrated that it is neither toxic nor mutagenic and activates macrophages. We now evaluate CA effects on cytokine production and gene expression from mice macrophages. The global view of changes in expression of genes with known functions can provide a vivid picture of the way in which cell adapts to a changing environment or a challenge. We found a decrease in IL-2 and IL-4 production and a differential expression in 147 genes from CA group. These genes are mainly involved in transcription/translation, cell structure and dynamics, immune response, cytoprotection, enzymatic process, and receptors/ligands. With gene expression analysis we state that this medication provokes a reaction that involves alterations in gene expression profile mainly in the ones involved with macrophages activation, corroborating the laboratorial research and the clinical data. J. Cell. Biochem. 104: 1364–1377, 2008. © 2008 Wiley-Liss, Inc.
Abbreviations used:
CA, Canova; Mϕ, Macrophages; HS, sucussed hydro-alcoholic solution; H, non-sucussed hydro-alcoholic solution; N, group without any treatment; DEGs, differently expressed genes; DH, decimal of Hahnemann's dilution.
Biological response modifiers are the most recent forms of immune modulatory therapy. They are aimed at specific cells or cytokines that contribute to the immune response [Ballow and Nelson, 1997]. Canova (CA) is a Brazilian medical formulation based on homeopathic techniques. It is an immunotherapeutic that stimulates the host's own defense favoring a particular immunologic response against malignant cells and microbial invasion. Monocytes/macrophages represent an important group of immune cells which play a central role in the inflammatory reaction to immune challenge producing pro-inflammatory cytokines, chemokines, reactive oxygen species (respiratory burst) and performing phagocytosis and antigen presentation. Previous studies demonstrated that CA activates macrophages (Mϕ) both in vivo and in vitro. In addition, tumor necrosis factor-α (TNFα) in vitro production is significantly diminished [Piemonte and Buchi, 2002]. NAD(P)H oxidase activity was increased as well as that of inducible nitric oxide synthase (iNOS), consequently producing reactive oxygen species (ROS) and nitric oxide (NO) respectively. NO inhibited cytochrome oxidase and peroxisomes activities [de Oliveira et al., 2006]. CA stimulates an increase of endosomal/lysosomal system, increases macrophages phagocytic activity of non-infective microorganisms (Saccharomyces cerevisiae and Trypanosoma cruzi epimastigotes) and diminishes phagocytic activity of infective microorganisms T. cruzi trypomastigotes and Leishmania amazonensis [Lopes et al., 2006]. CA modulatory effects were also observed in experimental infection both in vivo and in vitro by L. amazonensis, controlling progression of infection and limiting it dissemination [Pereira et al., 2005] and also in experimental infection with Paracoccidioides brasiliensis in a way that depends, at least in part, on NO production [Takahachi et al., 2006]. Sarcoma 180-bearing mice treated with CA showed reduction in sarcoma size and a significant infiltration of lymphoid cells, granulation tissue and fibrosis surrounding the tumor. CA enhanced T CD4 cells in the normal-treated group and increased both B and NK cells in S180-treated groups [Sato et al., 2005]. CA influence total bone marrow cells activating the monocytic lineage (CD11b) and the stromal cells (adherent cells) [Abud et al., 2006] as well as bone marrow progenitor cells leading to mononuclear cell differentiation and activation (Cesar et al., in press). Clinical reports showed successful results when CA is employed in diseases where the immune system is depressed due to its modulatory capacity [Sasaki et al., 2001]. Moreover, it is neither toxic nor mutagenic [Seligmann et al., 2003].
Mϕ play a significant role in the host defense mechanism [Forman and Torres, 2001]. Generally, macrophage activation is not a property of this cell type, but rather one that is acquired only following exposure to stimuli encountered in the microenvironment [Ohmori and Hamilton, 1994]. Their activation is based in cellular and ultrastructural changes in morphology and increased functional activities [Stadler and Weck, 1978]. These characteristics are results from the transcriptional profile of the cells and therefore the pattern of gene expression in a cell can provide detailed information about its state [DeRisi et al., 1997].
Microarrays and Gene Chips® (Affymetrix, Inc., CA) are microscopic arrays of immobilized nucleic acids. Their use extends to a wide range of analytical methods built around detection of sequence-specific nucleic acid hybridization [Epstein and Butow, 2000]. It is the technique now regarded as the most promising tool for large-scale gene expression analysis. It is used either for quantification of expression levels or for identification of differentially expressed genes (DEGs) between two mRNA preparations, which is important for medical and pharmaceutical studies [Sakai et al., 2000]. The use of gene expression profiling to globally address physiological/immunological questions has provided a significant amount of biologically relevant information in recent years and many global genome studies using different mammalian cell and tissue types have been reported.
The aim of the present study was to analyze the global effects of Canova treatment changes in gene expression in mice macrophages. The global view of changes in expression of genes with known functions provides a vivid picture of the way in which the cell adapts to a changing environment or challenge. Knowing when and where a gene is expressed often provides a strong clue as to its biological role [DeRisi et al., 1997].
MATERIALS AND METHODS
Canova (CA)
Canova is a commercial medicine that represents a new form of immunomodulatory therapy and follows Hahnemann's ancient homeopathic techniques. Canova is an aqueous, colorless, and odorless solution produced and sold in authorized Brazilian drugstores. Being a homeopathic medication, it was treated as so. One of homeopathy principles is that the effects of the substances become stronger the more they are diluted, and one important step in their preparation is called sucussion: “shakes” that are applied to the substance after each dilution stage. All homeopathic medications must be sucussed (vigorously shaken) in order to be activated. Thus Canova was always vigorously sucussed immediately before treatment.
Mother tinctures are purchased from authorized agencies indicated by the Brazilian Health Ministry. These agencies assure the quality (endotoxin free) and physico-chemical composition of its products. Starting from the original mother tincture (in the case of a plant this is an ethanolic extract) several dinamizations—sucussion (shaking) and dilution in distilled water—are performed. The Canova components are soluble in water. The final commercial product, Canova, is composed of 11 dH Aconitum napellus (Ranunculaceae), 19 dH Thuya occidentalis (Cupresaceae), 18 dH Bryonia alba (Curcubitaceae), 19 dH Arsenicum album (arsenic trioxide), 18 dH Lachesis muta (Viperidae) and less than 1% ethanol in distilled water. In our experiments we used the commercial product manipulated by Narciso da Lozzo Neto CRF-PR 5604.
Animals
Six- to 8-week-old male Swiss mice from the Central Animal House of the Universidade Federal do Paraná (UFPR) received a standard laboratory diet (Purina®) and water ad libitum. All recommendations of the National Law (No 6.638, November, 5th 1979) for scientific management of animals were respected and the Institutional Animal Care Committee at UFPR approved all related practices. Experiments were carried out at the Laboratório de Pesquisa em Células Neoplásicas e Inflamatórias, UFPR, which has a management program for produced residues.
CA Treatment
We used three controls group to compare the response of macrophages after mice treatment with Canova medication. All experiments were performed at least three times. The treatment groups were held according to a previous description [Piemonte and Buchi, 2002]. Mice were treated in vivo at daily intervals. They received seven doses (7 µl/g per day) of commercial CA injected subcutaneously. Canova was always vigorously sucussed immediately before treatment. Control mice were treated with a sucussed (HS) and non-sucussed (H) ethanolic solution (less than 1%) and mice without any treatment (N) were also used. The H group was only used in the GeneChip® assay.
Cell Preparation
Mϕ were washed out from mice peritoneal cavities with 10 ml of cold RNase free phosphate buffer solution (PBS) pH 7.4. Harvested peritoneal cells were counted in Neubauer chamber. All used materials and solutions were carefully held and protected to maintain the RNase free conditions.
Immunophenotyping
Specific antibodies were used for cell identification. Leukocytes were labeled using monoclonal antibodies (mAbs) anti-CD45. CD11b is a commonly used marker for the monocyte/macrophage lineage and the CD3 is mainly found in T lymphocytes [Lai et al., 1998]. Washed peritoneal cells (1 × 106) were incubated with 0.5 µg/ml phycoerythrin (PE) labeled CD45, CD11b, and CD3 antibodies in PBS for 40 min in the dark. After incubation, the cells were washed, resuspended in PBS and the fluorescence was analyzed according to standard procedures on a FACSCalibur flow cytometer (Becton Dickenson—BD), equipped with an argon ion laser (488 nm). Data were analyzed in Cell Quest program (BD). Data were submitted to analysis of variance (ANOVA) and Tukey test (P < 0.05) to determine the statistical significance of the intergroup comparisons. Data are representative of three independent experiments.
Determination of Cytokine Concentration
We also evaluated the capacity of these cells to elicit TNF-α, interferon-γ (IFN-γ) and interleukins 2, 4, and 5 (IL-2, IL-4, IL-5). After centrifugation, the peritoneal cells were discarded and the production of cytokines was measured in the supernatant by a Mouse Th1/Th2 cytokine CBA kit (BD—Pharmingen) according to the manufacturer's instructions. Fluorescence was measured by flow cytometry. The cytokine concentration was obtained by comparing data with a cytokine curve in the CBA program (BD). Data analysis was performed with ANOVA and Tukey test (P < 0.05).
RNA Isolation, cDNA Purification, and Biotinylated cRNA Synthesis
Total RNA was extracted from the cells using RNeasy mini Kits (QIAGEN), as described by the manufacturer. The extracted RNA concentration was measured by spectroscopy at 260 nm (nucleic acid optical density). Northern blot was used to verify the presence of RNA. cDNA was synthesized from the RNA of each group using an initiator oligonucleotide T7−(dT)24(5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′) and the reagents from Invitrogen. GeneChip® Sample Cleanup Module (Affymetrix, Inc.) was used for cDNA purification, as described by the manufacturer. The synthesis of biotinylated cRNA was performed with an in vitro transcription using the BioArray™ High Yield™ RNA Transcript Labeling Kit (Enzo Diagnostics) following the manufacturer's instructions. A suitable quantity of cRNA was fragmented and used for hybridization.
RNA Hybridization and Quantification
GeneChip® MG74Av2 (Affymetrix, Inc.) was used to characterize the changes in gene expression that take place during the process of Mϕ activation by Canova. The hybridization was performed according to the manufacturer's recommendations. GeneChip® Fluidics Station 400 was used to wash the chip and the probe arrays were scanned at GeneArray® Scanner (Affymetrix, Inc.) equipped with an argon ion laser, programmed following the manufacturer's instructions. The capture and initial analysis of hybridization images were performed with the software MicroArray Suite 5.0—MAS of Affymetrix, Inc. At the end of this process, a CEL file was created, containing the intensity value for each probe present in the GeneChip®, which was processed as follows.
Statistical Analysis of Differentially Expressed Genes (DEGs)
Data from the CEL file obtained from three independent experiments were analyzed by the Robust Multi-Array method—RMA [Irizarry et al., 2003]. A determination of differential expression was carried out by using the Significance Analysis of Microarray method—SAM [Tusher et al., 2001]. The criteria for the selection of DEGs were a fold change of two times and a false discovery rate of 5%. Data are representative of three independent experiments.
RESULTS
Immunophenotyping
Mouse cell surface antigens were used to identify peritoneal cell populations. Immunostaining was performed with a panel of antibodies to leukocytes (CD45) in general, T lymphocyte marker (CD3) and macrophages (CD11b). We found no statistical differences among the groups (CA, N, and HS). Approximately 95% of peritoneal cells were leukocytes. CD11b positive cells were the most abundant cells among them (84%). Almost 20% of total cells were CD3 positive. The results are shown in Figure 1.
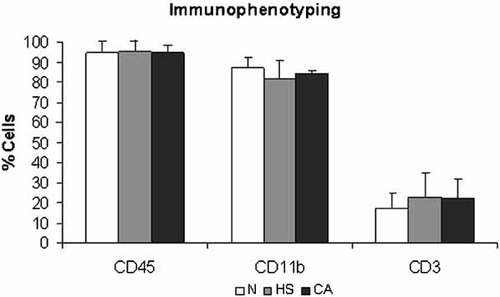
Immunophenotyping—specific cell surface antigens were used for cell identification and the fluorescence was analyzed by a flow cytometer. We found no statistical differences among the groups (CA, N, and HS). Pan-leukocyte marker (CD45) was found in approximately 95% of the cells. CD11b positive cells were the most abundant cells (84%). Almost 20% of total cells were CD3 positive. Results are expressed as mean ± SD. Data represent three independent experiments; *P < 0.05.
Determination of Cytokine Concentration
Supernatant cytokines were measured by a Mouse Th1/Th2 cytokine CBA kit in supernatant after centrifugation of recently extracted cells. A CBA program was used to calculate cytokine concentration. After statistical analyses we found a decrease in IL-4 and IL-2 in vivo production by cells from the CA group (Fig. 2). The HS group also presented a decrease in IL-2 production when compared to the N group. No statistical differences were found in TNFα, IFNγ, and IL-5 production (data not shown).
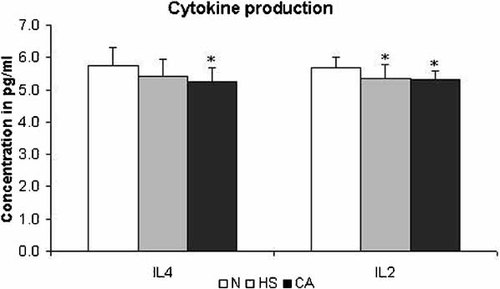
Cytokine determination—supernatant of peritoneal exudate was tested for IFN-γ, TNF-α, IL-2, IL-4, and IL-5 with mouse Th1/Th2 cytokine CBA kit. We observed a decrease in IL-4 and IL-2 in vivo production by the CA group. The HS group also presented a decrease in IL-2 production when compared to the N group. The other supernatant cytokines (TNF-α, IFN-γ, and IL-5) were not modified when this methodology was applied (data not shown). Results are expressed as mean ± SD (*P < 0.05). Data are representative of three independent experiments.
RNA Isolation—Hybridization
Rapidly after cell collection, total RNA was extracted from cells using RNeasy mini Kits (QIAGEN). The extracted RNA concentration was measured by spectroscopy at 260 nm. The RNA from each group was hybridized in GeneChips® MG74Av2 and analyzed. We found that the gene expression profile is modified in the CA group. Statistical analysis of microarray data revealed 147 genes differentially expressed after mice treatment.
CA Stimulation of Macrophages (Mϕ) Elicits Global Transcriptional Changes
The remarkable plasticity and diversity in the immune response derives from modulation of cells gene expression [Ohmori and Hamilton, 1994]. Relevant differences in gene expression were not observed among the groups HS, H, and N. Few differently expressed genes (DEGs) were found when we compared these groups with each other. They were mainly involved in cell cycle and signal transduction in the N group (data not shown). After data processing with RMA and SAM method, with FC and FDR thresholds of twofold and 5%, respectively, we found that the CA group differentially expressed 147 genes (45 were upregulated and 102 were downregulated) when compared with the HS group (Table IA and B). Plots were created to illustrate the comparisons (Fig. 3A between the control groups and Fig. 3B CA × HS comparison). We decided to use HS group as the control group, being that any difference found would be actually only the CA response.
| Fold change | Short form | |
|---|---|---|
| (A) 45 genes upregulated by CA | ||
| Cell signaling and signal transduction | ||
| 2.70 | Metallothionein 1 | Mt1 |
| 7.80 | Metallothionein 2 | Mt2 |
| 2.07 | Olfactory receptor 71 | Olfr71 |
| 5.49 | Suppressor of cytokine signaling 3 | Socs3 |
| 2.15 | Suppressor of cytokine signaling 3 | Socs3 |
| Cytoskeleton/cell adhesion | ||
| 2.34 | Actin, beta, cytoplasmic | Actb |
| 2.05 | Dynein, cytoplasmic, intermediate chain 1 | Dncic1 |
| 2.15 | Talin | Tln |
| 2.61 | Syndecan 4 | Sdc4 |
| Immune response | ||
| 2.42 | Histocompatibility 2, class II antigen E beta | H2-Eb1 |
| 4.57 | Heme oxygenase (decycling) 1 | Hmox1 |
| 5.60 | HLA-B-associated transcript 9 | Bat9 |
| Metabolism | ||
| 2.54 | Mevalonate kinase | Mvk |
| 2.70 | GM2 ganglioside activator protein | Gm2a |
| Transport | ||
| 4.67 | Adaptor protein complex AP-2, alpha 2 subunit | Ap2a2 |
| 6.18 | Hemoglobin alpha, adult chain 1 | Hba-a1 |
| 17.46 | Hemoglobin alpha, adult chain 1 | Hba-a1 |
| 3.92 | Hemoglobin, beta adult major chain | Hbb-b1 |
| 2.19 | Thioredoxin reductase 3 | Txnrd3 |
| Cell cycle | ||
| 3.60 | Growth arrest and DNA-damage-inducible 45 beta | Gadd45b |
| 2.15 | Talin | Tln |
| 2.19 | RIKEN cDNA 1700051C09 gene | 1700051C09Rik |
| Transcription | ||
| 2.55 | Ribosomal protein S28 | Rps28 |
| 6.46 | Small nuclear ribonucleoprotein B | Snrpb |
| 2.16 | Centaurin, gamma 3 | Centg3 |
| 7.14 | Heat shock protein 1A | Hspa1a |
| 5.60 | Heat shock protein 1B | Hspa1b |
| 4.66 | Splicing factor 3a, subunit 2 | Sf3a2 |
| 3.43 | Sequestosome 1 | Sqstm1 |
| Others | ||
| 2.23 | Fatty acid desaturase 1 | Fads1 |
| 3.03 | Mitochondrial ribosomal protein S18A | Mrps18a |
| 2.09 | Protease, serine, 25 | Prss25 |
| Unknown function | ||
| 2.30 | P53-variant (p53) | — |
| 2.25 | Aspartyl aminopeptidase | Dnpep |
| 2.14 | Hypothetical gene supported by BC030401 (LOC380819), mRNA | — |
| 3.14 | Zinc finger, CCHC domain containing 7 | Zcchc7 |
| 2.23 | Solute carrier family 39 (zinc transporter), member 4 | Slc39a4 |
| 4.65 | RIKEN cDNA 9430029L20 gene | 9430029L20Rik |
| 2.59 | RIKEN cDNA A430104N18 gene | A430104N18Rik |
| 3.92 | DNA segment, Chr 14, Wayne State University 89, expressed | D14Wsu89e |
| 2.12 | Ephrin A3 | Efna3 |
| 2.39 | Erythroid differentiation regulator 1 | Erdr1 |
| 2.01 | Expressed sequence R74740 | R74740 |
| 2.19 | RNA binding motif protein 9 | Rbm9 |
| 2.56 | Secretin | Sct |
| (B) 102 genes downregulated by CA | ||
| Cell signaling and signal transduction | ||
| 0.43 | Annexin A1 | Anxa1 |
| 0.29 | Expressed sequence AW111922 | AW111922 |
| 0.42 | Expressed sequence AW111922 | AW111922 |
| 0.43 | Protein tyrosine phosphatase, receptor type, O | Ptpro |
| 0.43 | RAB9, member RAS oncogene family | Rab9 |
| 0.38 | RAS p21 protein activator 4 | Rasa4 |
| Cytoskeleton/cell adhesion | ||
| 0.34 | Activated leukocyte cell adhesion molecule | Alcam |
| 0.39 | Integrin alpha M | Itgam |
| Immune response | ||
| 0.48 | Bone marrow stromal cell antigen 1 | Bst1 |
| 0.40 | Chemokine (C-C motif) ligand 5 | Ccl5 |
| 0.49 | Chemokine (C-C) receptor 2 | Ccr2 |
| 0.43 | CD1d1 antigen | Cd1d1 |
| 0.31 | Chemokine (C-X-C motif) receptor 4 | Cxcr4 |
| 0.49 | Dual specificity phosphatase 1 | Dusp1 |
| 0.45 | Guanylate nucleotide binding protein 1 | Gbp1 |
| 0.36 | Guanylate nucleotide binding protein 2 | Gbp2 |
| 0.37 | Histocompatibility 2, Q region locus 7 | H2-Q7 |
| 0.47 | Interferon inducible protein 1 | Ifi1 |
| 0.42 | Interferon gamma inducible protein | Ifi47 |
| 0.29 | Interferon activated gene 202B | Ifi202b |
| 0.36 | Interferon activated gene 205 | Ifi205 |
| 0.46 | Interferon gamma induced GTPase | Igtp |
| 0.50 | Proteosome (prosome, macropain) subunit, beta type 9 (large multifunctional protease 2) | Psmb9 |
| Metabolism | ||
| 0.46 | Aldehyde dehydrogenase 9, subfamily A1 | Aldh9a1 |
| 0.47 | Adenylosuccinate synthetase 2, non-muscle | Adss2 |
| 0.47 | Glucose-6-phosphate dehydrogenase X-linked | G6pdx |
| 0.41 | Glyoxalase 1 | Glo1 |
| 0.43 | Glycogenin 1 | Gyg1 |
| Transport | ||
| 0.45 | RIKEN cDNA 1110032D12 gene | 1110032D12Rik |
| 0.49 | ADP-ribosylation factor-like 6 interacting protein 5 | Arl6ip5 |
| 0.41 | ATPase, Ca++ transporting, ubiquitous | Atp2a3 |
| 0.50 | Chloride channel 3 | Clcn3 |
| 0.40 | Cytochrome b-245, beta polypeptide | Cybb |
| 0.49 | Electron transferring flavoprotein, alpha polypeptide | Etfa |
| 0.48 | Fatty acid binding protein 7, brain | Fabp7 |
| 0.49 | Sorting nexin 10 | Snx10 |
| 0.45 | Syntaxin 3 | Stx3 |
| 0.47 | Syntaxin binding protein 3 | Stxbp3 |
| 0.38 | Thioredoxin domain containing 7 | Txndc7 |
| 0.34 | Thioredoxin domain containing 7 | Txndc7 |
| Cell cycle | ||
| 0.38 | MAD2 (mitotic arrest deficient, homolog)-like 1 (yeast) | Mad2l1 |
| 0.41 | Antigen identified by monoclonal antibody Ki 67 | Mki67 |
| 0.36 | Schlafen 1 | Slfn1 |
| 0.49 | Schlafen 2 | Slfn2 |
| 0.46 | Schlafen 4 | Slfn4 |
| 0.49 | Transforming growth factor, beta 2 | Tgfb2 |
| Transcription | ||
| Sigla | ||
| 0.46 | RIKEN cDNA 1110054N06 gene | 1110054N06Rik |
| 0.36 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Cited2 |
| 0.40 | Cleavage stimulation factor, 3' pre-RNA subunit 2 | Cstf2 |
| 0.48 | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | Dhx36 |
| 0.47 | Eukaryotic translation initiation factor 3, subunit 6 | Eif3s6 |
| 0.46 | Fragile X mental retardation syndrome 1 homolog | Fmr1 |
| 0.43 | GA repeat binding protein, alpha | Gabpa |
| 0.47 | Pre-B-cell leukemia transcription factor 3 | Pbx3 |
| 0.50 | p300/CBP-associated factor | Pcaf |
| 0.37 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Smarca2 |
| 0.23 | Signal transducer and activator of transcription 1 | Stat1 |
| 0.44 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor | Taf9 |
| 0.39 | Transcription elongation factor A (SII) 1 | Tcea1 |
| 0.48 | Tripartite motif protein 30 | Trim30 |
| 0.37 | Wilms' tumor 1-associating protein | Wtap |
| 0.48 | Zinc finger protein 62 | Zfp62 |
| 0.47 | Zinc finger protein 265 | Zfp265 |
| Others | ||
| 0.44 | Formyl peptide receptor, related sequence 2 | Fpr-rs2 |
| 0.44 | Macrophage receptor with collagenous structure | Marco |
| 0.45 | Mitochondrial ribosomal protein L36 | Mrpl36 |
| 0.39 | X-linked myotubular myopathy gene 1 | Mtm1 |
| 0.44 | Numb gene homolog (Drosophila) | Numb |
| 0.44 | Pyruvate dehydrogenase kinase, isoenzyme 3 | Pdk3 |
| 0.40 | Quaking | Qk |
| 0.50 | Ribophorin II | Rpn2 |
| 0.35 | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | Sdha |
| 0.42 | Sestrin 1 | Sesn1 |
| 0.37 | Signal sequence receptor, gamma | Ssr3 |
| 0.48 | Striatin, calmodulin binding protein 3 | Strn3 |
| 0.32 | T-cell specific GTPase | Tgtp |
| 0.43 | Ubiquitin-conjugating enzyme E2E 1, UBC4/5 homolog (yeast) | Ube2e1 |
| 0.40 | Vesicle-associated membrane protein 3 | Vamp3 |
| Unknown function | ||
| 0.48 | — | — |
| 0.44 | Diabetic nephropathy-related gene 1 mRNA, partial sequence | — |
| 0.41 | Transcribed sequence with moderate similarity to protein sp:P00722 (E. coli) BGAL_ECOLI Beta-galactosidase | — |
| 0.41 | — | — |
| 0.49 | Transcribed sequences | — |
| 0.49 | RIKEN cDNA 0610010K14 gene | 0610010K14Rik |
| 0.44 | RIKEN cDNA 1110021E09 gene | 1110021E09Rik |
| 0.44 | RIKEN cDNA 2310015N07 gene | 2310015N07Rik |
| 0.45 | RIKEN cDNA 2610042L04 gene | 2610042L04Rik |
| 0.44 | RIKEN cDNA 2610205H19 gene | 2610205H19Rik |
| 0.47 | RIKEN cDNA 6330442E10 gene | 6330442E10Rik |
| 0.45 | RIKEN cDNA 9130022A11 gene | 9130022A11Rik |
| 0.49 | Expressed sequence AI265322 | AI265322 |
| 0.46 | Expressed sequence AI788669 | AI788669 |
| 0.44 | Expressed sequence AW112010 | AW112010 |
| 0.41 | DNA-damage-inducible transcript 4 | Ddit4 |
| 0.46 | Heterogeneous nuclear ribonucleoprotein H2 | Hnrph2 |
| 0.49 | Influenza virus NS1A binding protein | Ivns1abp |
| 0.37 | LIM domain only 7 | Lmo7 |
| 0.48 | Open reading frame 18 | ORF18 |
| 0.36 | Ring finger protein 138 | Rnf138 |
- These genes were selected after statistical analysis with SAM method. The DEGs observed are mainly involved in transcriptional and translational processes, cell structure and dynamics, immune response, cytoprotection, enzymatic process, chemokine receptors and their ligands.
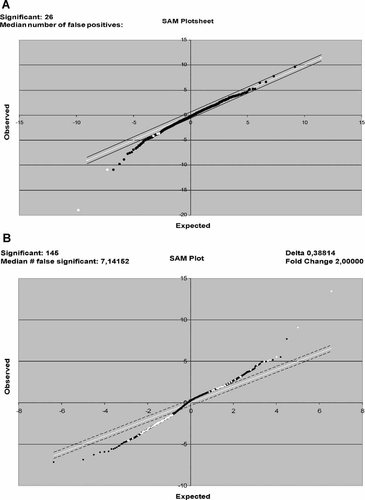
SAM plot displaying DEGs identified by the SAM method (with FC and FDR thresholds of twofold and 5%, respectively), after data processing with RMA. Graph A illustrates a comparison between the control groups (H × HS groups, as an example). Graph B shows the CA × HS comparison, where 145 DEGs were identified (45 were upregulated and 102 were downregulated, in relation to the CA group).
The DEGs observed are mainly involved in transcriptional and translational processes, cell structure and dynamics, immune response, cytoprotection, enzymatic process, chemokine receptors and their ligands. We describe here some of the DEGs observed in Mϕ treated with CA.
DISCUSSION
CA is being used in Brazil by patients with diseases where the immune system is depressed such as with acquired immunodeficiency syndrome (AIDS), hepatitis and neoplasia. It is a biological response modifier that acts on macrophages, thus enhancing the individual's own immunity to favor a particular immunologic response.
Macrophages (Mϕ) play an essential role in host defense against infection and tumoral cells. A large body of data indicates that macrophages must be activated in order to achieve efficiency [Ohmori and Hamilton, 1994]. Recent studies have described the effects of CA on Mϕ. About 86% of treated Mϕ were activated when observed in light microscopy and transmission electron microscopy [Piemonte and Buchi, 2002]. Now with immunophenotyping we were able to demonstrate that approximately 95% of peritoneal cells were leukocytes and within these Mϕ/monocytes cells were the most abundant. There were no statistical differences in cell percentages among the groups, showing that CA does not act quantitatively but qualitatively on Mϕ via their activation.
Mϕ are involved in many different process. When activated they can release products such as ROS and tumor necrosis factor that are direct harmful to microorganisms or cancer cells. On the other hand, they play an indirect role in these anti-microbial and anti-tumor activities by secretion of cytokines or by antigen processing and presentation, thereby regulating the immune system [Klimp et al., 2002]. We already found that CA treatment alters some of these Mϕ properties [Piemonte and Buchi, 2002; de Oliveira et al., 2006; Lopes et al., 2006]. We have now demonstrated that in vivo treatment with CA decreases the production of IL-2 and IL-4. Cote-Sierra et al. 2004 demonstrated that in vivo IL-2 neutralization inhibits Il-4 production in two models.
Interleukins are cytokines that influence the biological behavior either of the cells producing the cytokine or cell types of different lineages. They have a decisive role in determining the immune status, and the integrity of the immune system is controlled by efficient cellular responses to cytokine stimulation [Rees, 1992]. IL-4 plays an active role in regulating macrophage inflammatory activities. Zhou et al. 1995 demonstrated that it is an important negative regulator of macrophage. They described a decrease in superoxide anion production by macrophages due to suppression of NADPH oxidase activity by IL-4. We have found a decrease in IL-4 production and we demonstrated that CA increases NADPH oxidase activity [de Oliveira et al., 2006], fact that is in agreement with those authors. IL-2 is central to processes involved in the development of an antigen specific and antigen non-specific immune responses. It also influences the behavior of other cell types, and several reports have shown that IL-2 is an essential growth factor for the function of B cells, eosinophils, monocytes, and macrophages [Rees, 1992]. Sbarba et al. 1996 demonstrated that IL-2 down-modulates the macrophage colony-stimulating factor receptor (M-CSF.R) in murine macrophages. Macrophage colony-stimulating factor (M-CSF) is the main growth factor for mononuclear phagocytes, supporting its survival and stimulating its proliferation and differentiation. We found a decrease in IL-2 production by Mϕ. The effectiveness of the Mϕ population depends on the balance between their responses to proliferation or activation stimuli. These responses tend to be antagonistic, and the proliferation of activated cells is usually reduced. Thus on diminishing IL-2 in the cell environment the efficacy of Mϕ treated with CA tends to be higher assuming a specific functional state. We also observed a downregulation in IL-2 production by the HS group. We have observed that sucussed ethanolic solutions present some effects on Mϕ (data not published). Despite its effects on IL-2 production, changes in gene expression were not observed.
Macrophage activation is, among other criteria, characterized by cellular and ultrastructural changes in its morphology. Since many of these functional activities require increased protein synthesis, it is very likely that the cellular RNA content could be used as a parameter to identify cells with increased functional activities. Thus, analysis of gene expression in macrophages treated with CA can provide detailed information about its state and may contribute to understanding its way of action. In this assay RNA extracted from peritoneal exudate cells was mainly representative of Mϕ since it was found to be the majority of peritoneal cells in the immunophenotyping assay. We found that CA stimulated Mϕ to elicit global transcriptional changes. This treatment induced a robust up or downregulation of some genes observed by their fold change. Fold change analysis detect all and only genes that are differently expressed due to biologic variation [Irizarry et al., 2003]. For better understanding the action of CA on macrophages, we created some groups based on the gene function.
Transcriptional and Translational Processes
Many of the cellular changes that accompany Mϕ activation depend upon modulation of specific gene expression. In eukaryotes it is controlled at a number of levels including (1) transcriptional mechanisms, (2) post-transcriptional mechanisms, (3) translational efficiency, and (4) post-translational modifications [Ohmori and Hamilton, 1994]. We found an enhanced gene expression of proteins related to this processes. For instance, centaurin gamma 3 (Centg3) is a protein necessary to regulate transcription; snRNP B is a protein from spliceosomes that is involved in splicing [Kambach et al., 1999]; splicing factor 3a (SF3a) that function in 3′splice site recognition at early stages of spliceosome assembly [Tanackovic and Krämer, 2005]; ribosomal protein S28 and mitochondrial ribosomal protein S18A were increased in Mϕ treated with CA.
Changes in transcriptional and translational profiles are consequences of increased macrophage activities. Thus upregulation of genes involved with these activities was already anticipated.
Cell Structure and Dynamics
CA increased the production of molecules linked to extracellular and intracellular interactions.
Piemonte and Buchi in 2002 showed that macrophages treated with CA presented redistribution in fibronectin receptors (α5β1 integrin) and in cytoskeleton actin filaments. We found an upregulation on β actin expression, which is in accordance with the previous data. Talin, an actin binding protein [Brakebusch and Fässler, 2003], which is an important focal adhesion component was also increased. Another adhesion protein that was upregulated by CA is syndecan-4. It appears to be a coreceptor in adhesion to a range of extracellular ligands, modifying the primary integrin-mediated responses [Woods and Couchman, 2001].
We already have described that CA treatment enhanced the spreading ability of Mϕ and induced immunity favoring a particular immunologic response against microorganisms through the phagocytic pathway [Lopes et al., 2006]. Increasing the macrophage's capacity to deliver phagosomes into lysosomes is essential to phagocytosis efficiency. The bi-directional movement of phagosomes along microtubules is mediated by cytoplasmic dynein and kinesin [Rabinowitz et al., 1992; Blocker et al., 1997]. We found that dynein intermediate chain was upregulated by CA.
Macrophages with increased receptor expression may act better in signal transduction pathways. Ephrin A3 was found to be upregulated by CA. Eph receptors and ephrins activate signal transduction pathways via binding of a variety of cytoplasmic proteins that regulate adhesion and organization of the actin cytoskeleton [Poliakov and Cotrina Wilkinson, 2004].
Major histocompatibility complex class II (MHCII) molecules are essential to present peptides generated in the intracellular vesicles of macrophages to CD4+ T helper lymphocytes during the induction phase of immune responses [Yun et al., 1999]. CA increased the expression of histocompatibility class II antigen E beta. It seems that CA activates macrophages to enhance phagocytic capacity as well as to induce MHCII expression improving antigen presentation.
With these data we can state that CA is increasing cell attachment and adhesion as well as its functional consequences, such as phagocytosis and antigen presentation.
Immune Response
Several suppressors of cytokine signaling (SOCS) are key physiological regulators of the immune system. We find that the suppressor of cytokine signaling 3 (SOCS 3) was upregulated in Mϕ treated with CA. It a positive regulator of antigen presenting cells (APCs), is involved in T cell differentiation and also regulates T cell function by negatively regulating IL-2 signaling [Kubo et al., 2003]. Our observation of a decrease in IL-2 production by treated cells may corroborate the finding of SOCS3 induction.
CD1 belongs to a large group of non-classic molecules that are structurally related to the classical antigen-presenting MHC I molecules. In mice it is encoded by two genes, CD1d1 and CD1d2 [Bradbury et al., 1988]. We have found a CD1d1 downregulation by CA. CD1d1 deficiency leads to a strong reduction in the number of NT cells accompanied by a dramatic decrease in early IL-4 production [Mendiratta et al., 1997]. Once again, our observation of decreased production of IL-4 by treated cells may be in agreement with this gene repression.
Stress-Related Proteins: Cytoprotection
Macrophages triggered with CA had NAD(P)H oxidase activity increased as well as that of inducible nitric oxide synthase (iNOS), consequently producing the ROS and nitric oxide (NO) respectively [de Oliveira et al., 2006]. In order to protect cells and tissues from radical-induced toxicity, cells must have induction of the gene expression of scavengers [Adler et al., 1999]. We observed that some proteins that are often produced under stress conditions were upregulated like Hsp70-1 (1A and 1B), metallothioneins (MT-1 and MT-2), heme oxygenase-1 (HO-1), thioredoxin reductase (TR-3), growth arrest and DNA-damage-inducible 45 beta (GADD45β), erythroid differentiation regulator (EDR) and sequestosome (SQSTM1). Cellular antioxidants appear to be crucial for the reduction of oxidative stress [Poss and Tonegawa, 1997].
One interesting fact is that if Mϕ overexpress HO-1 the pro-inflammatory response (e.g., TNFα production) is markedly inhibited whereas the anti-inflammatory response (e.g., IL-10 production) is enhanced [Minamino et al., 2001]. Piemonte and Buchi 2002 described an in vitro decrease in TNFα production by CA, but we did not observe any change in in vivo TNFα production, probably due to Mϕ interaction with other cell types.
As we have already discussed, ROS are being produced by macrophages challenged with CA, so that many cytoprotective molecules are being differentially expressed. Transcriptional upregulation of pro-survival genes like GADD45β [De Smaele et al., 2001] is very important since CA activated Mϕ are producing many cytotoxic agents such ROS and NO.
These observations may corroborate the concept that CA is activating Mϕ oxidative metabolism and consequentially participating in redox signaling via upregulation of the cytoprotective gene.
Enzymes
CA increased mevalonate kinase (MK) gene expression. MK deficiency in inherited disorders leads to immune system dysregulation [Houten et al., 2000], so that augmenting this enzyme expression may have an important biological significance. CA also upregulated aspartyl aminopeptidase (Dnpep) expression. Dnpep is likely to play an important role in intracellular protein and peptide metabolism. Activated macrophages may have increased proteolysis and peptidolysis function to better accomplish antigen processing and presentation, thereby regulating the immune system. Serine protease (Prss25) activity was increased as well by CA and it is also involved in proteolysis and peptidolysis. Human immunodeficiency virus type-1 (HIV-1) reverse transcriptase (RT) plays a central role in the virus replication cycle. Degradation of HIV-1 RT by direct interaction with a host cell serine protease leads to a non-productive infection [Château et al., 2001]. Increasing serine protease levels in macrophages, CA may improve the immune response against invading viruses.
Another enzyme gene expression increased by CA was GM2 ganglioside activator protein (Gm2a). GM2a is required in pathways that generate lipid mediators that may control intracellular protein trafficking, vesicular transport, membrane ruffling, cell motility, mitogenesis, oncogenesis, and inflammation [Nakamura et al., 1998]. Fatty acid desaturase 1 (FADS1) was upregulated by CA. With the improvement of this enzyme function, CA is probably promoting high levels of fatty acid that influence a wide variety of cellular processes including vesicle fusion, protein export and modification, enzyme activation or deactivation, cell signaling, membrane permeability, bacterial pathogenesis, and transcriptional control [Black and DiRusso, 2003].
We found that the dual specificity phosphatase 1 (Dusp1) was downregulated by CA. It is known to inactivate in vitro mitogen-activated protein kinase. The MAP kinase 1,2/protein kinase C system is an intracellular signaling network that regulates many cellular machines [Alessi et al., 1993]. Thus, downregulating this gene, CA may have MAP kinase pathways available in any activation process where it is needed.
These enzymes discussed above are essential to the activation process of macrophages since they participate in various related process from cell morphology to biochemical reactions.
Chemokine Receptors and Ligands
The chemokines are a family of pro-inflammatory cytokines that attract and activate a variety of cells. Two main chemokine families have been identified, namely CC and CXC based on the position of the first two cysteine residues [Mellado et al., 2001]. The CC-chemokine RANTES (CCL5) along with the receptors CCR2 and CXCR4, were downregulated in cells treated with CA.
Numerous publications over the last decade have demonstrated the importance of chemokine receptors for the human immunodeficiency virus (HIV) entry. Two CC chemokine receptors, CCR3 and CCR5, as well as the CXCR4 chemokine receptor, have been shown to be necessary for infection by HIV-1 [Frade et al., 1997]. In fact, CCR5 and CXCR4 act synergistically with CD4 in an ordered multistep mechanism to allow the binding and entry of the HIV-1 virus [Singer et al., 2001]. Another CC chemokine receptor, CCR2 was found to act as a coreceptor for HIV infection [Frade et al., 1997]. The CC-chemokine RANTES (CCL5) can enhance HIV infection of target cells in a manner that is independent of CD4 and any known coreceptor, and even independent of the route of virus entry. It cross-links glycosaminoglycans, such as heparan sulfate, on both the virion and cell membrane [Trkola et al., 1999]. Receptors and ligand downregulation by CA may interfere with virus entry by reducing the density of available receptors and coreceptors on the cell surface interfering with propagation of infection. Recently, we observed an improvement in the life quality of HIV positive patients, with an increase in CD4 positive cells (data not published yet).
CA, as we have demonstrated, increases antioxidant defenses. Antioxidants can decrease expression of CCR2, CCR5, and CXCR4 [Saccani et al., 2000]. Thus the downregulation of these proteins may be explained by redox regulation induced by this medication.
Other Receptors
We have found a decrease in formyl peptide receptor related sequence-2 (Fpr-rs2) gene expression. Fpr mediates phagocyte chemotactic responses, increasing cell migration and the release of pro-inflammatory mediators [Le et al., 2001]. Annexin I and HIV-1 envelope proteins were found to contain domains that interact with FPR receptors [Le et al., 2001]. We have found as well that annexin A1 was downregulated by Canova. Annexin I is deployed by phagocytes as receptors for the recognition of apoptotic cells [Fan et al., 2004].
The macrophage receptor with collagenous structure (MARCO) belongs to the class A scavenger receptor molecules, and are able to bind various substances [Kraal et al., 2000]. CA decreased MARCO gene expression in mouse Mϕ.
The emergence of influenza A viruses highlights the need for antiviral drugs, particularly considering the potential threat of a pandemic of H5N1 influenza A viruses, the most lethal virus of bird flu. Until now, no medication was successfully developed against the bird flu due to the high virus mutability. We found that the CA treatment decreased the expression of influenza virus NS1A binding protein. This change must be further studied due to its relevance.
Whether the downregulation of these receptors have an influence on CA immunomodulator effects is unknown and requires further studies.
Genes Induced by IFN
Interferons are cytokines that have antiviral effects and inhibit tumor cell proliferation. They induce a large number of genes in their target cells. Although we found no alterations in IFN-γ production, we observed downregulation in some genes regulated by interferon. These are, interferon activated gene 205, 202A, interferon gamma inducible protein, interferon inducible protein 1, interferon gamma induced GTPase, and guanylate nucleotide binding proteins 1 and 2. These changes in gene expression occur for reasons we do not understand yet. Further studies will be necessary to answer these questions.
CONCLUSION
Immunotherapy is a form of medical treatment based upon the concept of modulating the immune system to achieve a therapeutic goal. The interest in this therapeutic modality has increased during the past decade. Advances in molecular immunology and gene therapy have been contributing to the development of new strategies to fight against immune-mediated disease as well as infections and tumor cells. Therefore, understanding how immune modulators act may lead to an essential strategy for the future success of immunotherapy. Screening thousands of genes at the same time, microarray analysis could offer new insight into the defense networks and could improve our understanding of the mechanism by which this medicine is beneficial in clinical practice.
With gene expression analysis, we are able to demonstrate that CA acts on Mϕ, triggering a reaction that involves alterations in gene expression profile, mainly in those involved with Mϕ activation. The genes found with this analysis raise many questions for further scientifically approaches. The understanding of how these are connected with each other and how they are linked to clinical findings must be further investigated. With the delineation of expressed genes we can propose and investigate the possible action pathways of Canova.
These results indicate a strong biochemical support of the clinical results found in the latest 50 years in patients with many different diseases. Unfortunately, the majority of medical doctors in South America did not publish the clinical findings. Therefore macrophage seems to be a potential therapeutic target for the regulation of immune responses by Canova medication.
Acknowledgements
The authors thank the Secretaria de Ciência e Tecnologia do Paraná, Brasil and CAPES for financial support. We also thank Dr. Philip A.J. Gorin for the careful revision of this manuscript.




